Transcripts synthesized by RNA polymerase III can be polyadenylated in an AAUAAA-dependent manner
- PMID: 18658125
- PMCID: PMC2525947
- DOI: 10.1261/rna.1006608
Transcripts synthesized by RNA polymerase III can be polyadenylated in an AAUAAA-dependent manner
Abstract
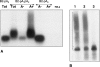
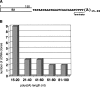
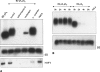
It is well known that nearly all eukaryotic mRNAs contain a 3' poly(A) tail. A polyadenylation signal (AAUAAA) nearby the 3' end of pre-mRNA is required for poly(A) synthesis. The protein complex involved in the pre-mRNA polyadenylation is coupled with RNA polymerase II during the transcription of a gene. According to the commonly accepted view, only RNAs synthesized by RNA polymerase II can be polyadenylated in an AAUAAA-dependent manner. Here we report the polyadenylation of short interspersed elements (SINEs) B2 and VES transcripts generated by RNA polymerase III. HeLa cells were transfected with SINE constructs with or without polyadenylation signals. The analyses of the SINE transcripts showed that only the RNAs with the AAUAAA-signal contained poly(A) tails. Polyadenylated B2 RNA was found to be much more stable in cells than B2 RNA without a poly(A) tail.
Figures




Similar articles
-
[Polyadenylation of Sine Transcripts Generated by RNA Polymerase III Dramatically Prolongs Their Lifetime in Cells].Mol Biol (Mosk). 2020 Jan-Feb;54(1):78-86. doi: 10.31857/S0026898419040165. Mol Biol (Mosk). 2020. PMID: 32163391 Russian.
-
Identification of nucleotide sequences and some proteins involved in polyadenylation of RNA transcribed by Pol III from SINEs.RNA Biol. 2021 Oct;18(10):1475-1488. doi: 10.1080/15476286.2020.1857942. Epub 2020 Dec 11. RNA Biol. 2021. PMID: 33258402 Free PMC article.
-
Polyadenylation of RNA transcribed from mammalian SINEs by RNA polymerase III: Complex requirements for nucleotide sequences.Biochim Biophys Acta. 2016 Feb;1859(2):355-65. doi: 10.1016/j.bbagrm.2015.12.003. Epub 2015 Dec 14. Biochim Biophys Acta. 2016. PMID: 26700565
-
[Canonical and noncanonical RNA polyadenylation].Mol Biol (Mosk). 2017 Mar-Apr;51(2):262-273. doi: 10.7868/S0026898417010189. Mol Biol (Mosk). 2017. PMID: 28537233 Review. Russian.
-
Reexamining the polyadenylation signal: were we wrong about AAUAAA?Mol Cell Endocrinol. 2002 Apr 25;190(1-2):1-8. doi: 10.1016/s0303-7207(02)00044-8. Mol Cell Endocrinol. 2002. PMID: 11997173 Review.
Cited by
-
Crosstalk between vault RNAs and innate immunity.Mol Biol Rep. 2024 Mar 5;51(1):387. doi: 10.1007/s11033-024-09305-y. Mol Biol Rep. 2024. PMID: 38443657 Free PMC article. Review.
-
Novel viral splicing events and open reading frames revealed by long-read direct RNA sequencing of adenovirus transcripts.PLoS Pathog. 2022 Sep 12;18(9):e1010797. doi: 10.1371/journal.ppat.1010797. eCollection 2022 Sep. PLoS Pathog. 2022. PMID: 36095031 Free PMC article.
-
Ere, a Family of Short Interspersed Elements in the Genomes of Odd-Toed Ungulates (Perissodactyla).Animals (Basel). 2024 Jul 5;14(13):1982. doi: 10.3390/ani14131982. Animals (Basel). 2024. PMID: 38998094 Free PMC article.
-
Origin and evolution of SINEs in eukaryotic genomes.Heredity (Edinb). 2011 Dec;107(6):487-95. doi: 10.1038/hdy.2011.43. Epub 2011 Jun 15. Heredity (Edinb). 2011. PMID: 21673742 Free PMC article. Review.
-
SINEs as Potential Expression Cassettes: Impact of Deletions and Insertions on Polyadenylation and Lifetime of B2 and Ves SINE Transcripts Generated by RNA Polymerase III.Int J Mol Sci. 2023 Sep 27;24(19):14600. doi: 10.3390/ijms241914600. Int J Mol Sci. 2023. PMID: 37834047 Free PMC article.
References
-
- Anderson J.T. RNA turnover: Unexpected consequences of being tailed. Curr. Biol. 2005;15:R635–R638. - PubMed
-
- Bentley D.L. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. - PubMed
-
- Bernstein P., Ross J. Poly(A), poly(A) binding protein and the regulation of mRNA stability. Trends Biochem. Sci. 1989;14:373–377. - PubMed
-
- Borodulina O.R., Kramerov D.A. Wide distribution of short interspersed elements among eukaryotic genomes. FEBS Lett. 1999;457:409–413. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
