Molecular dynamic simulations analysis of ritonavir and lopinavir as SARS-CoV 3CL(pro) inhibitors
- PMID: 18706430
- PMCID: PMC7094092
- DOI: 10.1016/j.jtbi.2008.07.030
Molecular dynamic simulations analysis of ritonavir and lopinavir as SARS-CoV 3CL(pro) inhibitors
Abstract
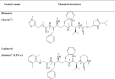
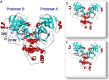
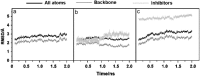
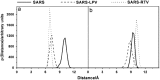
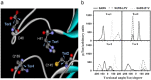
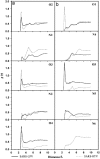
Since the emergence of the severe acute respiratory syndrome (SARS) to date, neither an effective antiviral drug nor a vaccine against SARS is available. However, it was found that a mixture of two HIV-1 proteinase inhibitors, lopinavir and ritonavir, exhibited some signs of effectiveness against the SARS virus. To understand the fine details of the molecular interactions between these proteinase inhibitors and the SARS virus via complexation, molecular dynamics simulations were carried out for the SARS-CoV 3CL(pro) free enzyme (free SARS) and its complexes with lopinavir (SARS-LPV) and ritonavir (SARS-RTV). The results show that flap closing was clearly observed when the inhibitors bind to the active site of SARS-CoV 3CL(pro). The binding affinities of LPV and RTV to SARS-CoV 3CL(pro) do not show any significant difference. In addition, six hydrogen bonds were detected in the SARS-LPV system, while seven hydrogen bonds were found in SARS-RTV complex.
Figures
Similar articles
-
Why Are Lopinavir and Ritonavir Effective against the Newly Emerged Coronavirus 2019? Atomistic Insights into the Inhibitory Mechanisms.Biochemistry. 2020 May 12;59(18):1769-1779. doi: 10.1021/acs.biochem.0c00160. Epub 2020 Apr 24. Biochemistry. 2020. PMID: 32293875
-
Exploring the Binding Mechanism of PF-07321332 SARS-CoV-2 Protease Inhibitor through Molecular Dynamics and Binding Free Energy Simulations.Int J Mol Sci. 2021 Aug 24;22(17):9124. doi: 10.3390/ijms22179124. Int J Mol Sci. 2021. PMID: 34502033 Free PMC article.
-
Molecular Docking of Azithromycin, Ritonavir, Lopinavir, Oseltamivir, Ivermectin and Heparin Interacting with Coronavirus Disease 2019 Main and Severe Acute Respiratory Syndrome Coronavirus-2 3C-Like Proteases.J Nanosci Nanotechnol. 2021 Apr 1;21(4):2075-2089. doi: 10.1166/jnn.2021.19029. J Nanosci Nanotechnol. 2021. PMID: 33500022
-
Lopinavir/ritonavir: a protease inhibitor for HIV-1 treatment.Expert Opin Pharmacother. 2008 Sep;9(13):2363-75. doi: 10.1517/14656566.9.13.2363. Expert Opin Pharmacother. 2008. PMID: 18710360 Review.
-
Lopinavir/ritonavir: Repurposing an old drug for HIV infection in COVID-19 treatment.Biomed J. 2021 Mar;44(1):43-53. doi: 10.1016/j.bj.2020.11.005. Epub 2020 Nov 10. Biomed J. 2021. PMID: 33608241 Free PMC article. Review.
Cited by
-
Identification of steroidal cardenolides from Calotropis procera as novel HIV-1 PR inhibitors: A molecular docking & molecular dynamics simulation study.Indian J Med Res. 2024 Jul;160(1):78-86. doi: 10.25259/IJMR_2115_23. Indian J Med Res. 2024. PMID: 39382500 Free PMC article.
-
Current and future therapeutical approaches for COVID-19.Drug Discov Today. 2020 Aug;25(8):1545-1552. doi: 10.1016/j.drudis.2020.06.018. Epub 2020 Jun 20. Drug Discov Today. 2020. PMID: 32574697 Free PMC article. Review.
-
Therapeutic _targeting of Innate Immune Receptors Against SARS-CoV-2 Infection.Front Pharmacol. 2022 Jun 30;13:915565. doi: 10.3389/fphar.2022.915565. eCollection 2022. Front Pharmacol. 2022. PMID: 35847031 Free PMC article. Review.
-
Lung tissue distribution of drugs as a key factor for COVID-19 treatment.Br J Pharmacol. 2020 Nov;177(21):4995-4996. doi: 10.1111/bph.15102. Epub 2020 Jun 9. Br J Pharmacol. 2020. PMID: 32424836 Free PMC article. No abstract available.
-
Review of trials currently testing treatment and prevention of COVID-19.Clin Microbiol Infect. 2020 Aug;26(8):988-998. doi: 10.1016/j.cmi.2020.05.019. Epub 2020 May 23. Clin Microbiol Infect. 2020. PMID: 32454187 Free PMC article. Review.
References
-
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main protease (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. - PubMed
-
- Carlson H.A. Protein flexibility and drug design: how to hit a moving _target. Curr. Opin. Chem. Biol. 2002;6:447–452. - PubMed
-
- Carlson H.A., McCammon J.A. Accommodating protein flexibility in computational drug design. Mol. Pharmacol. 2000;57:213–218. - PubMed
-
- Case D.A., Pearlman J.W., Caldwell T.E., Cheatham J., Jr., Wang W.S., Ross C.L., Simmerling T.A., Darden K.M., Merz R.V., Stanton A.L., Cheng J.J., Vincent M., Crowley V., Tsui H., Gohlke R.J., Radmer Y., Duan J., Pitera I., Massova G.L., Seibel U.C., Singh P.K., Kollman P.A. University of California; San Francisco, CA: 2002. AMBER. (Version 7.0 ed.)
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

