In vivo reconstitution of autophagy in Saccharomyces cerevisiae
- PMID: 18725539
- PMCID: PMC2518709
- DOI: 10.1083/jcb.200801035
In vivo reconstitution of autophagy in Saccharomyces cerevisiae
Abstract
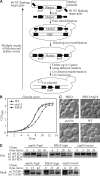
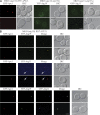
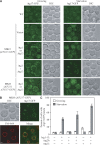
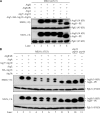
Autophagy is a major intracellular degradative pathway that is involved in various human diseases. The role of autophagy, however, is complex; although the process is generally considered to be cytoprotective, it can also contribute to cellular dysfunction and disease progression. Much progress has been made in our understanding of autophagy, aided in large part by the identification of the autophagy-related (ATG) genes. Nonetheless, our understanding of the molecular mechanism remains limited. In this study, we generated a Saccharomyces cerevisiae multiple-knockout strain with 24 ATG genes deleted, and we used it to carry out an in vivo reconstitution of the autophagy pathway. We determined minimum requirements for different aspects of autophagy and studied the initial protein assembly steps at the phagophore assembly site. In vivo reconstitution enables the study of autophagy within the context of the complex regulatory networks that control this process, an analysis that is not possible with an in vitro system.
Figures

Similar articles
-
New insights into autophagy using a multiple knockout strain.Autophagy. 2008 Nov;4(8):1073-5. doi: 10.4161/auto.6962. Epub 2008 Nov 10. Autophagy. 2008. PMID: 18971623 Free PMC article.
-
Monitoring the Formation of Autophagosomal Precursor Structures in Yeast Saccharomyces cerevisiae.Methods Enzymol. 2017;588:323-365. doi: 10.1016/bs.mie.2016.09.085. Epub 2016 Nov 21. Methods Enzymol. 2017. PMID: 28237109
-
Atg27 is a second transmembrane cycling protein.Autophagy. 2007 May-Jun;3(3):254-6. doi: 10.4161/auto.3823. Epub 2007 May 11. Autophagy. 2007. PMID: 17297289
-
Mechanistic Insights into the Role of Atg11 in Selective Autophagy.J Mol Biol. 2020 Jan 3;432(1):104-122. doi: 10.1016/j.jmb.2019.06.017. Epub 2019 Jun 22. J Mol Biol. 2020. PMID: 31238043 Free PMC article. Review.
-
Atg9 trafficking in the yeast Saccharomyces cerevisiae.Autophagy. 2007 Mar-Apr;3(2):145-8. doi: 10.4161/auto.3608. Epub 2007 Mar 21. Autophagy. 2007. PMID: 17204846 Review.
Cited by
-
Autophagic processes in yeast: mechanism, machinery and regulation.Genetics. 2013 Jun;194(2):341-61. doi: 10.1534/genetics.112.149013. Genetics. 2013. PMID: 23733851 Free PMC article. Review.
-
The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy.J Cell Biol. 2010 Jan 11;188(1):101-14. doi: 10.1083/jcb.200904075. J Cell Biol. 2010. PMID: 20065092 Free PMC article.
-
The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy.Nature. 2013 Aug 22;500(7463):468-71. doi: 10.1038/nature12313. Epub 2013 Jul 17. Nature. 2013. PMID: 23863932 Free PMC article.
-
Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy.Proc Natl Acad Sci U S A. 2010 Apr 27;107(17):7811-6. doi: 10.1073/pnas.1000063107. Epub 2010 Apr 7. Proc Natl Acad Sci U S A. 2010. PMID: 20375281 Free PMC article.
-
Atg35, a micropexophagy-specific protein that regulates micropexophagic apparatus formation in Pichia pastoris.Autophagy. 2011 Apr;7(4):375-85. doi: 10.4161/auto.7.4.14369. Epub 2011 Apr 1. Autophagy. 2011. PMID: 21169734 Free PMC article.
References
-
- Bowers, K., and T.H. Stevens. 2005. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1744:438–454. - PubMed
-
- Cao, Y., and D.J. Klionsky. 2007. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 17:839–849. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

