Chemical probes for histone-modifying enzymes
- PMID: 18800048
- PMCID: PMC2908280
- DOI: 10.1038/nchembio.111
Chemical probes for histone-modifying enzymes
Abstract
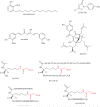
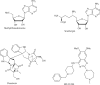
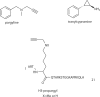
The histone-modifying enzymes that catalyze reversible lysine acetylation and methylation are central to the epigenetic regulation of chromatin remodeling. From the early discovery of histone deacetylase inhibitors to the more recent identification of histone demethylase blockers, chemical approaches offer increasingly sophisticated tools for the investigation of the structure and function of these lysine-modifying enzymes. This review summarizes progress to date on compounds identified from screens or by design that can modulate the activity of classical histone deacetylases, sirtuins, histone acetyltransferases, histone methyltransferases and histone demethylases. We highlight applications of compounds to mechanistic and functional studies involving these enzymes and discuss future challenges regarding _target specificity and general utility.
Figures
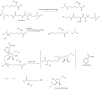
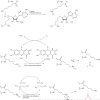
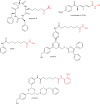
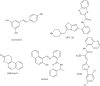
Similar articles
-
Small molecule control of chromatin remodeling.Chem Biol. 2014 Sep 18;21(9):1196-210. doi: 10.1016/j.chembiol.2014.07.024. Chem Biol. 2014. PMID: 25237863 Review.
-
Histone modification enzymes: novel _targets for cancer drugs.Expert Opin Emerg Drugs. 2004 May;9(1):135-54. doi: 10.1517/eoed.9.1.135.32947. Expert Opin Emerg Drugs. 2004. PMID: 15155140 Review.
-
Small molecule modulators of histone acetylation and methylation: a disease perspective.Biochim Biophys Acta. 2010 Oct-Dec;1799(10-12):810-28. doi: 10.1016/j.bbagrm.2010.09.005. Epub 2010 Oct 1. Biochim Biophys Acta. 2010. PMID: 20888936 Review.
-
Erasers of histone acetylation: the histone deacetylase enzymes.Cold Spring Harb Perspect Biol. 2014 Apr 1;6(4):a018713. doi: 10.1101/cshperspect.a018713. Cold Spring Harb Perspect Biol. 2014. PMID: 24691964 Free PMC article. Review.
-
Screening for compounds that modulate epigenetic regulation of the transcriptome: an overview.J Biomol Screen. 2011 Dec;16(10):1137-52. doi: 10.1177/1087057111417871. Epub 2011 Oct 14. J Biomol Screen. 2011. PMID: 22002420 Review.
Cited by
-
High-throughput screening-compatible assays of As(III) S-adenosylmethionine methyltransferase activity.Anal Biochem. 2015 Jul 1;480:67-73. doi: 10.1016/j.ab.2015.04.011. Epub 2015 Apr 10. Anal Biochem. 2015. PMID: 25866076 Free PMC article.
-
Lysine-Specific Demethylase 1 Inhibitors: A Comprehensive Review Utilizing Computer-Aided Drug Design Technologies.Molecules. 2024 Jan 22;29(2):550. doi: 10.3390/molecules29020550. Molecules. 2024. PMID: 38276629 Free PMC article. Review.
-
Peptide arrays identify isoform-selective substrates for profiling endogenous lysine deacetylase activity.ACS Chem Biol. 2010 Sep 17;5(9):863-73. doi: 10.1021/cb100088g. ACS Chem Biol. 2010. PMID: 20849068 Free PMC article.
-
Discovery of potent and selective histone deacetylase inhibitors via focused combinatorial libraries of cyclic alpha3beta-tetrapeptides.J Med Chem. 2009 Dec 10;52(23):7836-46. doi: 10.1021/jm900850t. J Med Chem. 2009. PMID: 19705846 Free PMC article.
-
BRD4 interacts with PML/RARα in acute promyelocytic leukemia.Front Med. 2018 Dec;12(6):726-734. doi: 10.1007/s11684-017-0604-x. Epub 2018 Dec 14. Front Med. 2018. PMID: 30552662
References
-
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. - PubMed
-
- Murray K. The occurrence of ε-N-methyl lysine in histones. Biochemistry. 1964;3:10–15. - PubMed
-
- Gershey EL, Vidali G, Allfrey VG. Chemical studies of histone acetylation. The occurrence of ε-N-acetyllysine in the f2a1 histone. J. Biol. Chem. 1968;243:5018–5022. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

