Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy
- PMID: 18829864
- PMCID: PMC2592676
- DOI: 10.1091/mbc.e08-05-0544
Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy
Abstract
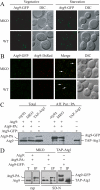
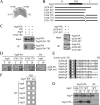
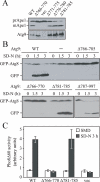
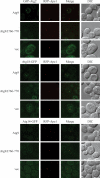
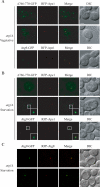
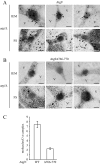
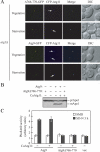
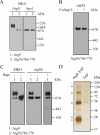
Autophagy is the degradation of a cell's own components within lysosomes (or the analogous yeast vacuole), and its malfunction contributes to a variety of human diseases. Atg9 is the sole integral membrane protein required in formation of the initial sequestering compartment, the phagophore, and is proposed to play a key role in membrane transport; the phagophore presumably expands by vesicular addition to form a complete autophagosome. It is not clear through what mechanism Atg9 functions at the phagophore assembly site (PAS). Here we report that Atg9 molecules self-associate independently of other known autophagy proteins in both nutrient-rich and starvation conditions. Mutational analyses reveal that self-interaction is critical for anterograde transport of Atg9 to the PAS. The ability of Atg9 to self-interact is required for both selective and nonselective autophagy at the step of phagophore expansion at the PAS. Our results support a model in which Atg9 multimerization facilitates membrane flow to the PAS for phagophore formation.
Figures
Similar articles
-
Atg9 trafficking in autophagy-related pathways.Autophagy. 2007 May-Jun;3(3):271-4. doi: 10.4161/auto.3912. Epub 2007 May 29. Autophagy. 2007. PMID: 17329962
-
Double duty of Atg9 self-association in autophagosome biogenesis.Autophagy. 2009 Apr;5(3):385-7. doi: 10.4161/auto.5.3.7699. Epub 2009 Apr 23. Autophagy. 2009. PMID: 19182520 Free PMC article.
-
Phosphorylation of Atg9 regulates movement to the phagophore assembly site and the rate of autophagosome formation.Autophagy. 2016;12(4):648-58. doi: 10.1080/15548627.2016.1157237. Autophagy. 2016. PMID: 27050455 Free PMC article.
-
Transcriptional regulation of ATG9 by the Pho23-Rpd3 complex modulates the frequency of autophagosome formation.Autophagy. 2014 Sep;10(9):1681-2. doi: 10.4161/auto.29641. Epub 2014 Jul 7. Autophagy. 2014. PMID: 25046109 Free PMC article. Review.
-
Atg9 trafficking in the yeast Saccharomyces cerevisiae.Autophagy. 2007 Mar-Apr;3(2):145-8. doi: 10.4161/auto.3608. Epub 2007 Mar 21. Autophagy. 2007. PMID: 17204846 Review.
Cited by
-
The Mucosal Immune System and Its Regulation by Autophagy.Front Immunol. 2016 Jun 22;7:240. doi: 10.3389/fimmu.2016.00240. eCollection 2016. Front Immunol. 2016. PMID: 27446072 Free PMC article. Review.
-
Bulk and selective autophagy cooperate to remodel a fungal proteome in response to changing nutrient availability.bioRxiv [Preprint]. 2024 Sep 26:2024.09.24.614842. doi: 10.1101/2024.09.24.614842. bioRxiv. 2024. PMID: 39386609 Free PMC article. Preprint.
-
Autophagy regulation through Atg9 traffic.J Cell Biol. 2012 Jul 23;198(2):151-3. doi: 10.1083/jcb.201206119. J Cell Biol. 2012. PMID: 22826119 Free PMC article.
-
Molecular Functions of Glycoconjugates in Autophagy.J Mol Biol. 2016 Aug 14;428(16):3305-3324. doi: 10.1016/j.jmb.2016.06.011. Epub 2016 Jun 23. J Mol Biol. 2016. PMID: 27345664 Free PMC article. Review.
-
Evolutionary conservation and metabolic significance of autophagy in algae.Philos Trans R Soc Lond B Biol Sci. 2024 Nov 18;379(1914):20230368. doi: 10.1098/rstb.2023.0368. Epub 2024 Sep 30. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 39343016 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

