H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome
- PMID: 19047520
- PMCID: PMC2652204
- DOI: 10.1101/gr.080861.108
H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome
Abstract
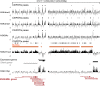
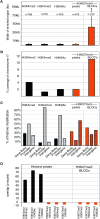
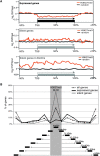
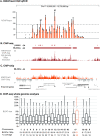
In mammals, genome-wide chromatin maps and immunofluorescence studies show that broad domains of repressive histone modifications are present on pericentromeric and telomeric repeats and on the inactive X chromosome. However, only a few autosomal loci such as silent Hox gene clusters have been shown to lie in broad domains of repressive histone modifications. Here we present a ChIP-chip analysis of the repressive H3K27me3 histone modification along chr 17 in mouse embryonic fibroblast cells using an algorithm named broad local enrichments (BLOCs), which allows the identification of broad regions of histone modifications. Our results, confirmed by BLOC analysis of a whole genome ChIP-seq data set, show that the majority of H3K27me3 modifications form BLOCs rather than focal peaks. H3K27me3 BLOCs modify silent genes of all types, plus flanking intergenic regions and their distribution indicates a negative correlation between H3K27me3 and transcription. However, we also found that some nontranscribed gene-poor regions lack H3K27me3. We therefore performed a low-resolution analysis of whole mouse chr 17, which revealed that H3K27me3 is enriched in mega-base-pair-sized domains that are also enriched for genes, short interspersed elements (SINEs) and active histone modifications. These genic H3K27me3 domains alternate with similar-sized gene-poor domains. These are deficient in active histone modifications, as well as H3K27me3, but are enriched for long interspersed elements (LINEs) and long-terminal repeat (LTR) transposons and H3K9me3 and H4K20me3. Thus, an autosome can be seen to contain alternating chromatin bands that predominantly separate genes from one retrotransposon class, which could offer unique domains for the specific regulation of genes or the silencing of autonomous retrotransposons.
Figures


Similar articles
-
Histone chaperone CAF-1 mediates repressive histone modifications to protect preimplantation mouse embryos from endogenous retrotransposons.Proc Natl Acad Sci U S A. 2015 Nov 24;112(47):14641-6. doi: 10.1073/pnas.1512775112. Epub 2015 Nov 6. Proc Natl Acad Sci U S A. 2015. PMID: 26546670 Free PMC article.
-
H4K20me3 co-localizes with activating histone modifications at transcriptionally dynamic regions in embryonic stem cells.BMC Genomics. 2018 Jul 3;19(1):514. doi: 10.1186/s12864-018-4886-4. BMC Genomics. 2018. PMID: 29969988 Free PMC article.
-
Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome.Mol Cell. 2007 Aug 3;27(3):353-66. doi: 10.1016/j.molcel.2007.06.024. Mol Cell. 2007. PMID: 17679087 Free PMC article.
-
The many faces of histone lysine methylation.Curr Opin Cell Biol. 2002 Jun;14(3):286-98. doi: 10.1016/s0955-0674(02)00335-6. Curr Opin Cell Biol. 2002. PMID: 12067650 Review.
-
Broad Chromatin Domains: An Important Facet of Genome Regulation.Bioessays. 2017 Dec;39(12). doi: 10.1002/bies.201700124. Epub 2017 Oct 23. Bioessays. 2017. PMID: 29058338 Review.
Cited by
-
Hepatic ontogeny and tissue distribution of mRNAs of epigenetic modifiers in mice using RNA-sequencing.Epigenetics. 2012 Aug;7(8):914-29. doi: 10.4161/epi.21113. Epub 2012 Jul 9. Epigenetics. 2012. PMID: 22772165 Free PMC article.
-
Chromatin landscape: methylation beyond transcription.Epigenetics. 2011 Jan;6(1):9-15. doi: 10.4161/epi.6.1.13331. Epub 2011 Jan 1. Epigenetics. 2011. PMID: 20855937 Free PMC article. Review.
-
Genome-wide prediction and analysis of human chromatin boundary elements.Nucleic Acids Res. 2012 Jan;40(2):511-29. doi: 10.1093/nar/gkr750. Epub 2011 Sep 19. Nucleic Acids Res. 2012. PMID: 21930510 Free PMC article.
-
The Role of Gammaherpesviruses in Cancer Pathogenesis.Pathogens. 2016 Feb 6;5(1):18. doi: 10.3390/pathogens5010018. Pathogens. 2016. PMID: 26861404 Free PMC article. Review.
-
H3K9me3-Dependent Heterochromatin: Barrier to Cell Fate Changes.Trends Genet. 2016 Jan;32(1):29-41. doi: 10.1016/j.tig.2015.11.001. Epub 2015 Dec 8. Trends Genet. 2016. PMID: 26675384 Free PMC article. Review.
References
-
- Azuara V., Perry P., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. - PubMed
-
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006a;125:315–326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
