Interactions between human glutamate carboxypeptidase II and urea-based inhibitors: structural characterization
- PMID: 19053759
- PMCID: PMC5516903
- DOI: 10.1021/jm800765e
Interactions between human glutamate carboxypeptidase II and urea-based inhibitors: structural characterization
Abstract
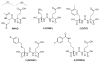
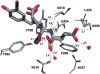
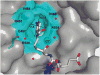
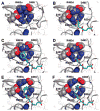
Urea-based, low molecular weight ligands of glutamate carboxypeptidase II (GCPII) have demonstrated efficacy in various models of neurological disorders and can serve as imaging agents for prostate cancer. To enhance further development of such compounds, we determined X-ray structures of four complexes between human GCPII and urea-based inhibitors at high resolution. All ligands demonstrate an invariant glutarate moiety within the S1' pocket of the enzyme. The ureido linkage between P1 and P1' inhibitor sites interacts with the active-site Zn(1)(2+) ion and the side chains of Tyr552 and His553. Interactions within the S1 pocket are defined primarily by a network of hydrogen bonds between the P1 carboxylate group of the inhibitors and the side chains of Arg534, Arg536, and Asn519. Importantly, we have identified a hydrophobic pocket accessory to the S1 site that can be exploited for structure-based design of novel GCPII inhibitors with increased lipophilicity.
Figures

Similar articles
-
Structural basis of interactions between human glutamate carboxypeptidase II and its substrate analogs.J Mol Biol. 2008 Mar 7;376(5):1438-50. doi: 10.1016/j.jmb.2007.12.066. Epub 2008 Jan 5. J Mol Biol. 2008. PMID: 18234225 Free PMC article.
-
Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.Bioorg Med Chem Lett. 2014 May 15;24(10):2340-5. doi: 10.1016/j.bmcl.2014.03.066. Epub 2014 Mar 28. Bioorg Med Chem Lett. 2014. PMID: 24731280 Free PMC article.
-
Design of composite inhibitors _targeting glutamate carboxypeptidase II: the importance of effector functionalities.FEBS J. 2016 Jan;283(1):130-43. doi: 10.1111/febs.13557. Epub 2015 Nov 5. FEBS J. 2016. PMID: 26460595 Free PMC article.
-
Glutamate carboxypeptidase II: an overview of structural studies and their importance for structure-based drug design and deciphering the reaction mechanism of the enzyme.Curr Med Chem. 2012;19(9):1300-9. doi: 10.2174/092986712799462667. Curr Med Chem. 2012. PMID: 22304708 Review.
-
Structure-activity relationships of glutamate carboxypeptidase II (GCPII) inhibitors.Curr Med Chem. 2012;19(9):1282-94. doi: 10.2174/092986712799462658. Curr Med Chem. 2012. PMID: 22304717 Review.
Cited by
-
Identification of alternative protein _targets of glutamate-ureido-lysine associated with PSMA tracer uptake in prostate cancer cells.Proc Natl Acad Sci U S A. 2022 Jan 25;119(4):e2025710119. doi: 10.1073/pnas.2025710119. Proc Natl Acad Sci U S A. 2022. PMID: 35064078 Free PMC article.
-
Synthesis and Preclinical Evaluation of a Bispecific PSMA-617/RM2 Heterodimer _targeting Prostate Cancer.ACS Med Chem Lett. 2024 Oct 18;15(11):1970-1978. doi: 10.1021/acsmedchemlett.4c00324. eCollection 2024 Nov 14. ACS Med Chem Lett. 2024. PMID: 39563828 Free PMC article.
-
A low molecular weight PSMA-based fluorescent imaging agent for cancer.Biochem Biophys Res Commun. 2009 Dec 18;390(3):624-9. doi: 10.1016/j.bbrc.2009.10.017. Epub 2009 Oct 8. Biochem Biophys Res Commun. 2009. PMID: 19818734 Free PMC article.
-
68Ga-labeled inhibitors of prostate-specific membrane antigen (PSMA) for imaging prostate cancer.J Med Chem. 2010 Jul 22;53(14):5333-41. doi: 10.1021/jm100623e. J Med Chem. 2010. PMID: 20568777 Free PMC article.
-
A Structure-Activity Relationship Study of Bimodal BODIPY-Labeled PSMA-_targeting Bioconjugates.ChemMedChem. 2021 Aug 19;16(16):2535-2545. doi: 10.1002/cmdc.202100210. Epub 2021 May 24. ChemMedChem. 2021. PMID: 33905162 Free PMC article.
References
-
- Sacha P, Zamecnik J, Barinka C, Hlouchova K, Vicha A, Mlcochova P, Hilgert I, Eckschlager T, Konvalinka J. Expression of glutamate carboxypeptidase II in human brain. Neuroscience. 2007;144:1361–1372. - PubMed
-
- Neale JH, Bzdega T, Wroblewska B. N-Acetylaspartylglutamate: the most abundant peptide neurotransmitter in the mammalian central nervous system. J Neurochem. 2000;75:443–452. - PubMed
-
- Riedel G, Platt B, Micheau J. Glutamate receptor function in learning and memory. Behav Brain Res. 2003;140:1–47. - PubMed
-
- Doble A. The role of excitotoxicity in neurodegenerative disease: implications for therapy. Pharmacol Ther. 1999;81:163–221. - PubMed
-
- Meldrum BS. Glutamate as a neurotransmitter in the brain: review of physiology and pathology. J Nutr. 2000;130:1007S–1015S. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
- CA1114111/CA/NCI NIH HHS/United States
- CA111982/CA/NCI NIH HHS/United States
- R21 MH080580-02/MH/NIMH NIH HHS/United States
- R21 MH080580/MH/NIMH NIH HHS/United States
- R21 CA111982-02/CA/NCI NIH HHS/United States
- R21 CA114111-02/CA/NCI NIH HHS/United States
- EB005423/EB/NIBIB NIH HHS/United States
- R33 MH080580/MH/NIMH NIH HHS/United States
- CA92871/CA/NCI NIH HHS/United States
- R21 EB005324-02/EB/NIBIB NIH HHS/United States
- R21 CA114111/CA/NCI NIH HHS/United States
- MH080580/MH/NIMH NIH HHS/United States
- R24 CA092871/CA/NCI NIH HHS/United States
- R21 CA111982/CA/NCI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- U24 CA092871/CA/NCI NIH HHS/United States
- R21 EB005324/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

