The unfolded protein response signals through high-order assembly of Ire1
- PMID: 19079236
- PMCID: PMC2846394
- DOI: 10.1038/nature07661
The unfolded protein response signals through high-order assembly of Ire1
Abstract
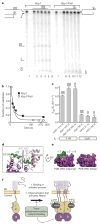
Aberrant folding of proteins in the endoplasmic reticulum activates the bifunctional transmembrane kinase/endoribonuclease Ire1. Ire1 excises an intron from HAC1 messenger RNA in yeasts and Xbp1 messenger RNA in metozoans encoding homologous transcription factors. This non-conventional mRNA splicing event initiates the unfolded protein response, a transcriptional program that relieves the endoplasmic reticulum stress. Here we show that oligomerization is central to Ire1 function and is an intrinsic attribute of its cytosolic domains. We obtained the 3.2-A crystal structure of the oligomer of the Ire1 cytosolic domains in complex with a kinase inhibitor that acts as a potent activator of the Ire1 RNase. The structure reveals a rod-shaped assembly that has no known precedence among kinases. This assembly positions the kinase domain for trans-autophosphorylation, orders the RNase domain, and creates an interaction surface for binding of the mRNA substrate. Activation of Ire1 through oligomerization expands the mechanistic repertoire of kinase-based signalling receptors.
Figures
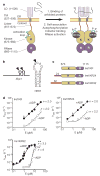
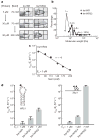
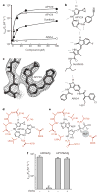

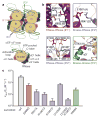
Comment in
-
Cell biology: How to combat stress.Nature. 2009 Feb 5;457(7230):668-9. doi: 10.1038/457668a. Nature. 2009. PMID: 19194438 No abstract available.
Similar articles
-
Specificity in endoplasmic reticulum-stress signaling in yeast entails a step-wise engagement of HAC1 mRNA to clusters of the stress sensor Ire1.Elife. 2014 Dec 30;3:e05031. doi: 10.7554/eLife.05031. Elife. 2014. PMID: 25549299 Free PMC article.
-
Bypassing a kinase activity with an ATP-competitive drug.Science. 2003 Nov 28;302(5650):1533-7. doi: 10.1126/science.1090031. Epub 2003 Oct 16. Science. 2003. PMID: 14564015
-
Conserved RNA structures in the non-canonical Hac1/Xbp1 intron.RNA Biol. 2011 Jul-Aug;8(4):552-6. doi: 10.4161/rna.8.4.15396. Epub 2011 Jul 1. RNA Biol. 2011. PMID: 21593604 Free PMC article.
-
Translation Control of HAC1 by Regulation of Splicing in Saccharomyces cerevisiae.Int J Mol Sci. 2019 Jun 12;20(12):2860. doi: 10.3390/ijms20122860. Int J Mol Sci. 2019. PMID: 31212749 Free PMC article. Review.
-
How IRE1 reacts to ER stress.Cell. 2008 Jan 11;132(1):24-6. doi: 10.1016/j.cell.2007.12.017. Cell. 2008. PMID: 18191217 Review.
Cited by
-
The unpredictability of prolonged activation of stress response pathways.J Cell Biol. 2015 Jun 22;209(6):781-7. doi: 10.1083/jcb.201503107. J Cell Biol. 2015. PMID: 26101215 Free PMC article.
-
Structural and Functional Analysis of the Allosteric Inhibition of IRE1α with ATP-Competitive Ligands.ACS Chem Biol. 2016 Aug 19;11(8):2195-205. doi: 10.1021/acschembio.5b00940. Epub 2016 Jun 9. ACS Chem Biol. 2016. PMID: 27227314 Free PMC article.
-
New insights on human IRE1 tetramer structures based on molecular modeling.Sci Rep. 2020 Oct 15;10(1):17490. doi: 10.1038/s41598-020-74347-8. Sci Rep. 2020. PMID: 33060689 Free PMC article.
-
XBP1S associates with RUNX2 and regulates chondrocyte hypertrophy.J Biol Chem. 2012 Oct 5;287(41):34500-13. doi: 10.1074/jbc.M112.385922. Epub 2012 Aug 3. J Biol Chem. 2012. Retraction in: J Biol Chem. 2015 Apr 24;290(17):10643. doi: 10.1074/jbc.A112.385922 PMID: 22865880 Free PMC article. Retracted.
-
Frustration in Fuzzy Protein Complexes Leads to Interaction Versatility.J Phys Chem B. 2021 Mar 18;125(10):2513-2520. doi: 10.1021/acs.jpcb.0c11068. Epub 2021 Mar 5. J Phys Chem B. 2021. PMID: 33667107 Free PMC article.
References
-
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. - PubMed
-
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. - PubMed
-
- Koong AC, Chauhan V, Romero-Ramirez L. _targeting XBP-1 as a novel anti-cancer strategy. Cancer Biol Ther. 2006;5:756–759. - PubMed
-
- Ma Y, Hendershot LM. The role of the unfolded protein response in tumour development: friend or foe? Nature Rev Cancer. 2004;4:966–977. - PubMed
-
- Zheng Y, et al. Hepatitis C virus non-structural protein NS4B can modulate an unfolded protein response. J Microbiol. 2005;43:529–536. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

