Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells
- PMID: 19147588
- PMCID: PMC2629396
- DOI: 10.1158/0008-5472.CAN-08-3415
Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells
Abstract
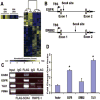
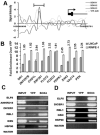
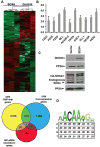
SOX4 is a critical developmental transcription factor in vertebrates and is required for precise differentiation and proliferation in multiple tissues. In addition, SOX4 is overexpressed in many human malignancies, but the exact role of SOX4 in cancer progression is not well understood. Here, we have identified the direct transcriptional _targets of SOX4 using a combination of genome-wide localization chromatin immunoprecipitation-chip analysis and transient overexpression followed by expression profiling in a prostate cancer model cell line. We have also used protein-binding microarrays to derive a novel SOX4-specific position-weight matrix and determined that SOX4 binding sites are enriched in SOX4-bound promoter regions. Direct transcriptional _targets of SOX4 include several key cellular regulators, such as EGFR, HSP70, Tenascin C, Frizzled-5, Patched-1, and Delta-like 1. We also show that SOX4 _targets 23 transcription factors, such as MLL, FOXA1, ZNF281, and NKX3-1. In addition, SOX4 directly regulates expression of three components of the RNA-induced silencing complex, namely Dicer, Argonaute 1, and RNA Helicase A. These data provide new insights into how SOX4 affects developmental signaling pathways and how these changes may influence cancer progression via regulation of gene networks involved in microRNA processing, transcriptional regulation, the TGFbeta, Wnt, Hedgehog, and Notch pathways, growth factor signaling, and tumor metastasis.
Figures

Similar articles
-
SOX4 interacts with plakoglobin in a Wnt3a-dependent manner in prostate cancer cells.BMC Cell Biol. 2011 Nov 19;12:50. doi: 10.1186/1471-2121-12-50. BMC Cell Biol. 2011. PMID: 22098624 Free PMC article.
-
SOX4 interacts with EZH2 and HDAC3 to suppress microRNA-31 in invasive esophageal cancer cells.Mol Cancer. 2015 Feb 3;14:24. doi: 10.1186/s12943-014-0284-y. Mol Cancer. 2015. PMID: 25644061 Free PMC article.
-
The SOX4/miR-17-92/RB1 Axis Promotes Prostate Cancer Progression.Neoplasia. 2019 Aug;21(8):765-776. doi: 10.1016/j.neo.2019.05.007. Epub 2019 Jun 22. Neoplasia. 2019. PMID: 31238254 Free PMC article.
-
SOX4: The unappreciated oncogene.Semin Cancer Biol. 2020 Dec;67(Pt 1):57-64. doi: 10.1016/j.semcancer.2019.08.027. Epub 2019 Aug 21. Semin Cancer Biol. 2020. PMID: 31445218 Free PMC article. Review.
-
The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe?Oncogene. 2013 Jul 18;32(29):3397-409. doi: 10.1038/onc.2012.506. Epub 2012 Dec 17. Oncogene. 2013. PMID: 23246969 Review.
Cited by
-
Annotation of functional variation in personal genomes using RegulomeDB.Genome Res. 2012 Sep;22(9):1790-7. doi: 10.1101/gr.137323.112. Genome Res. 2012. PMID: 22955989 Free PMC article.
-
Clinicopathological significance of SOX4 expression in primary gallbladder carcinoma.Diagn Pathol. 2012 Apr 17;7:41. doi: 10.1186/1746-1596-7-41. Diagn Pathol. 2012. PMID: 22510499 Free PMC article.
-
MicroRNA-129-5p modulates epithelial-to-mesenchymal transition by _targeting SIP1 and SOX4 during peritoneal dialysis.Lab Invest. 2015 Jul;95(7):817-832. doi: 10.1038/labinvest.2015.57. Epub 2015 May 11. Lab Invest. 2015. PMID: 25961171 Free PMC article.
-
Palate morphogenesis: current understanding and future directions.Birth Defects Res C Embryo Today. 2010 Jun;90(2):133-54. doi: 10.1002/bdrc.20180. Birth Defects Res C Embryo Today. 2010. PMID: 20544696 Free PMC article. Review.
-
CUL4B promotes prostate cancer progression by forming positive feedback loop with SOX4.Oncogenesis. 2019 Mar 14;8(3):23. doi: 10.1038/s41389-019-0131-5. Oncogenesis. 2019. PMID: 30872583 Free PMC article.
References
-
- Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. - PubMed
-
- Ya J, Schilham MW, de Boer PA, Moorman AF, Clevers H, Lamers WH. Sox4-deficiency syndrome in mice is an animal model for common trunk. Circ Res. 1998;83(10):986–94. - PubMed
-
- Wilson ME, Yang KY, Kalousova A, et al. The HMG box transcription factor Sox4 contributes to the development of the endocrine pancreas. Diabetes. 2005;54(12):3402–9. - PubMed
-
- Nissen-Meyer LS, Jemtland R, Gautvik VT, et al. Osteopenia, decreased bone formation and impaired osteoblast development in Sox4 heterozygous mice. J Cell Sci. 2007;120(Pt 16):2785–95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

