Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes
- PMID: 19287394
- PMCID: PMC2664327
- DOI: 10.1038/nmeth.1311
Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes
Abstract
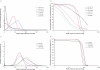
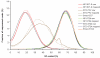
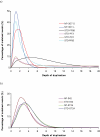
Amplification artifacts introduced during library preparation for the Illumina Genome Analyzer increase the likelihood that an appreciable proportion of these sequences will be duplicates and cause an uneven distribution of read coverage across the _targeted sequencing regions. As a consequence, these unfavorable features result in difficulties in genome assembly and variation analysis from the short reads, particularly when the sequences are from genomes with base compositions at the extremes of high or low G+C content. Here we present an amplification-free method of library preparation, in which the cluster amplification step, rather than the PCR, enriches for fully ligated template strands, reducing the incidence of duplicate sequences, improving read mapping and single nucleotide polymorphism calling and aiding de novo assembly. We illustrate this by generating and analyzing DNA sequences from extremely (G+C)-poor (Plasmodium falciparum), (G+C)-neutral (Escherichia coli) and (G+C)-rich (Bordetella pertussis) genomes.
Figures

Similar articles
-
Optimizing Illumina next-generation sequencing library preparation for extremely AT-biased genomes.BMC Genomics. 2012 Jan 3;13:1. doi: 10.1186/1471-2164-13-1. BMC Genomics. 2012. PMID: 22214261 Free PMC article.
-
Amplification-free library preparation for paired-end Illumina sequencing.Methods Mol Biol. 2011;733:257-66. doi: 10.1007/978-1-61779-089-8_18. Methods Mol Biol. 2011. PMID: 21431776
-
Sequencing of high G+C microbial genomes using the ultrafast pyrosequencing technology.J Biotechnol. 2011 Aug 20;155(1):68-77. doi: 10.1016/j.jbiotec.2011.04.010. Epub 2011 Apr 23. J Biotechnol. 2011. PMID: 21536083
-
De novo sequencing of plant genomes using second-generation technologies.Brief Bioinform. 2009 Nov;10(6):609-18. doi: 10.1093/bib/bbp039. Brief Bioinform. 2009. PMID: 19933209 Review.
-
Application of hybridization techniques to genome mapping and sequencing.Trends Genet. 1994 Mar;10(3):79-83. doi: 10.1016/0168-9525(94)90229-1. Trends Genet. 1994. PMID: 8178368 Review.
Cited by
-
Detection of ultra-rare mutations by next-generation sequencing.Proc Natl Acad Sci U S A. 2012 Sep 4;109(36):14508-13. doi: 10.1073/pnas.1208715109. Epub 2012 Aug 1. Proc Natl Acad Sci U S A. 2012. PMID: 22853953 Free PMC article.
-
Complete telomere-to-telomere de novo assembly of the Plasmodium falciparum genome through long-read (>11 kb), single molecule, real-time sequencing.DNA Res. 2016 Aug;23(4):339-51. doi: 10.1093/dnares/dsw022. Epub 2016 Jun 26. DNA Res. 2016. PMID: 27345719 Free PMC article.
-
Exome Sequencing: Current and Future Perspectives.G3 (Bethesda). 2015 Jul 2;5(8):1543-50. doi: 10.1534/g3.115.018564. G3 (Bethesda). 2015. PMID: 26139844 Free PMC article. No abstract available.
-
Identifying the causes and consequences of assembly gaps using a multiplatform genome assembly of a bird-of-paradise.Mol Ecol Resour. 2021 Jan;21(1):263-286. doi: 10.1111/1755-0998.13252. Epub 2020 Oct 10. Mol Ecol Resour. 2021. PMID: 32937018 Free PMC article.
-
A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica).Nat Genet. 2013 Aug;45(8):957-61. doi: 10.1038/ng.2673. Epub 2013 Jun 23. Nat Genet. 2013. PMID: 23793027
References
-
- Goman M, et al. The establishment of genomic DNA libraries for the human malaria parasite Plasmodium falciparum and identification of individual clones by hybridisation. Mol Biochem Parasitol. 1982;5:391–400. - PubMed
-
- Camargo AA, Fischer K, Lanzer M, del Portillo HA. Construction and characterization of a Plasmodium vivax genomic library in yeast artificial chromosomes. Genomics. 1997;42:467–473. - PubMed
-
- de Bruin D, Lanzer M, Ravetch JV. Characterization of yeast artificial chromosomes from Plasmodium falciparum: construction of a stable, representative library and cloning of telomeric DNA fragments. Genomics. 1992;14:332–339. - PubMed
-
- Triglia T, Kemp DJ. Large fragments of Plasmodium falciparum DNA can be stable when cloned in yeast artificial chromosomes. Mol Biochem Parasitol. 1991;44:207–211. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

