Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae
- PMID: 19302372
- PMCID: PMC2802268
- DOI: 10.1111/j.1474-9726.2009.00469.x
Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae
Abstract
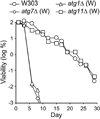
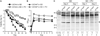
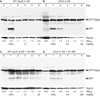
Following cessation of growth, yeast cells remain viable in a nondividing state for a period of time known as the chronological lifespan (CLS). Autophagy is a degradative process responsible for amino acid recycling in response to nitrogen starvation and amino acid limitation. We have investigated the role of autophagy during chronological aging of yeast grown in glucose minimal media containing different supplemental essential and nonessential amino acids. Deletion of ATG1 or ATG7, both of which are required for autophagy, reduced CLS, whereas deletion of ATG11, which is required for selective _targeting of cellular components to the vacuole for degradation, did not reduce CLS. The nonessential amino acids isoleucine and valine, and the essential amino acid leucine, extended CLS in autophagy-deficient as well as autophagy-competent yeast. This extension was suppressed by constitutive expression of GCN4, which encodes a transcriptional regulator of general amino acid control (GAAC). Consistent with this, GCN4 expression was reduced by isoleucine and valine. Furthermore, elimination of the leucine requirement extended CLS and prevented the effects of constitutive expression of GCN4. Interestingly, deletion of LEU3, a GAAC _target gene encoding a transcriptional regulator of branched side chain amino acid synthesis, dramatically increased CLS in the absence of amino acid supplements. In general, this indicates that activation of GAAC reduces CLS whereas suppression of GAAC extends CLS in minimal medium. These findings demonstrate important roles for autophagy and amino acid homeostasis in determining CLS in yeast.
Figures










Similar articles
-
Autophagy and leucine promote chronological longevity and respiration proficiency during calorie restriction in yeast.Exp Gerontol. 2013 Oct;48(10):1107-19. doi: 10.1016/j.exger.2013.01.006. Epub 2013 Jan 18. Exp Gerontol. 2013. PMID: 23337777 Free PMC article.
-
Amino acid homeostasis and chronological longevity in Saccharomyces cerevisiae.Subcell Biochem. 2012;57:161-86. doi: 10.1007/978-94-007-2561-4_8. Subcell Biochem. 2012. PMID: 22094422 Free PMC article. Review.
-
Gln3-Gcn4 hybrid transcriptional activator determines catabolic and biosynthetic gene expression in the yeast Saccharomyces cerevisiae.Biochem Biophys Res Commun. 2011 Jan 21;404(3):859-64. doi: 10.1016/j.bbrc.2010.12.075. Epub 2010 Dec 22. Biochem Biophys Res Commun. 2011. PMID: 21184740
-
SKO1 deficiency extends chronological lifespan in Saccharomyces cerevisiae.Biosci Biotechnol Biochem. 2019 Aug;83(8):1473-1476. doi: 10.1080/09168451.2019.1571901. Epub 2019 Jan 24. Biosci Biotechnol Biochem. 2019. PMID: 30676285
-
Is Gcn4-induced autophagy the ultimate downstream mechanism by which hormesis extends yeast replicative lifespan?Curr Genet. 2019 Jun;65(3):717-720. doi: 10.1007/s00294-019-00936-4. Epub 2019 Jan 23. Curr Genet. 2019. PMID: 30673825 Free PMC article. Review.
Cited by
-
Autophagy in ageing and ageing-associated diseases.Acta Pharmacol Sin. 2013 May;34(5):605-11. doi: 10.1038/aps.2012.188. Epub 2013 Feb 18. Acta Pharmacol Sin. 2013. PMID: 23416930 Free PMC article. Review.
-
Mechanisms of amino acid-mediated lifespan extension in Caenorhabditis elegans.BMC Genet. 2015 Feb 3;16(1):8. doi: 10.1186/s12863-015-0167-2. BMC Genet. 2015. PMID: 25643626 Free PMC article.
-
Hot topics in aging research: protein translation, 2009.Aging Cell. 2009 Dec;8(6):617-23. doi: 10.1111/j.1474-9726.2009.00522.x. Epub 2009 Sep 11. Aging Cell. 2009. PMID: 19747234 Free PMC article. Review.
-
Regulation of metabolic health by essential dietary amino acids.Mech Ageing Dev. 2019 Jan;177:186-200. doi: 10.1016/j.mad.2018.07.004. Epub 2018 Jul 22. Mech Ageing Dev. 2019. PMID: 30044947 Free PMC article. Review.
-
Diversification of Paralogous α-Isopropylmalate Synthases by Modulation of Feedback Control and Hetero-Oligomerization in Saccharomyces cerevisiae.Eukaryot Cell. 2015 Jun;14(6):564-77. doi: 10.1128/EC.00033-15. Epub 2015 Apr 3. Eukaryot Cell. 2015. PMID: 25841022 Free PMC article.
References
-
- Amberg DC, Burke DJ, Strathern JN. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. p. 230.
-
- Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–287. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

