High-throughput quantitative analysis of the human intestinal microbiota with a phylogenetic microarray
- PMID: 19363078
- PMCID: PMC2687317
- DOI: 10.1128/AEM.02764-08
High-throughput quantitative analysis of the human intestinal microbiota with a phylogenetic microarray
Abstract
Gut microbiota carry out key functions in health and participate in the pathogenesis of a growing number of diseases. The aim of this study was to develop a custom microarray that is able to identify hundreds of intestinal bacterial species. We used the Entrez nucleotide database to compile a data set of bacterial 16S rRNA gene sequences isolated from human intestinal and fecal samples. Identified sequences were clustered into separate phylospecies groups. Representative sequences from each phylospecies were used to develop a microbiota microarray based on the Affymetrix GeneChip platform. The designed microbiota array contains probes to 775 different bacterial phylospecies. In our validation experiments, the array correctly identified genomic DNA from all 15 bacterial species used. Microbiota array has a detection sensitivity of at least 1 pg of genomic DNA and can detect bacteria present at a 0.00025% level of overall sample. Using the developed microarray, fecal samples from two healthy children and two healthy adults were analyzed for bacterial presence. Between 227 and 232 species were detected in fecal samples from children, whereas 191 to 208 species were found in adult stools. The majority of identified phylospecies belonged to the classes Clostridia and Bacteroidetes. The microarray revealed putative differences between the gut microbiota of healthy children and adults: fecal samples from adults had more Clostridia and less Bacteroidetes and Proteobacteria than those from children. A number of other putative differences were found at the genus level.
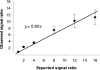
Figures
Similar articles
-
The human gut chip "HuGChip", an explorative phylogenetic microarray for determining gut microbiome diversity at family level.PLoS One. 2013 May 17;8(5):e62544. doi: 10.1371/journal.pone.0062544. Print 2013. PLoS One. 2013. PMID: 23690942 Free PMC article.
-
Dysbiosis of fecal microbiota in Crohn's disease patients as revealed by a custom phylogenetic microarray.Inflamm Bowel Dis. 2010 Dec;16(12):2034-42. doi: 10.1002/ibd.21319. Inflamm Bowel Dis. 2010. PMID: 20848492
-
Design and evaluation of oligonucleotide-microarray method for the detection of human intestinal bacteria in fecal samples.FEMS Microbiol Lett. 2002 Aug 6;213(2):175-82. doi: 10.1111/j.1574-6968.2002.tb11302.x. FEMS Microbiol Lett. 2002. PMID: 12167534
-
Optimizing the analysis of human intestinal microbiota with phylogenetic microarray.FEMS Microbiol Ecol. 2011 Feb;75(2):332-42. doi: 10.1111/j.1574-6941.2010.01009.x. Epub 2010 Dec 13. FEMS Microbiol Ecol. 2011. PMID: 21155851 Free PMC article.
-
Application of phylogenetic microarrays to interrogation of human microbiota.FEMS Microbiol Ecol. 2012 Jan;79(1):2-11. doi: 10.1111/j.1574-6941.2011.01222.x. Epub 2011 Nov 9. FEMS Microbiol Ecol. 2012. PMID: 22092522 Free PMC article. Review.
Cited by
-
DNA microarrays for the diagnosis of infectious diseases.Med Mal Infect. 2012 Oct;42(10):453-9. doi: 10.1016/j.medmal.2012.07.017. Epub 2012 Oct 9. Med Mal Infect. 2012. PMID: 23058632 Free PMC article. Review.
-
Is adolescence the missing developmental link in Microbiome-Gut-Brain axis communication?Dev Psychobiol. 2019 Jul;61(5):783-795. doi: 10.1002/dev.21821. Epub 2019 Jan 28. Dev Psychobiol. 2019. PMID: 30690712 Free PMC article. Review.
-
Functional metagenomic investigations of the human intestinal microbiota.Front Microbiol. 2011 Oct 17;2:188. doi: 10.3389/fmicb.2011.00188. eCollection 2011. Front Microbiol. 2011. PMID: 22022321 Free PMC article.
-
Species and genus level resolution analysis of gut microbiota in Clostridium difficile patients following fecal microbiota transplantation.Microbiome. 2014 Apr 21;2:13. doi: 10.1186/2049-2618-2-13. eCollection 2014. Microbiome. 2014. PMID: 24855561 Free PMC article.
-
The human gut chip "HuGChip", an explorative phylogenetic microarray for determining gut microbiome diversity at family level.PLoS One. 2013 May 17;8(5):e62544. doi: 10.1371/journal.pone.0062544. Print 2013. PLoS One. 2013. PMID: 23690942 Free PMC article.
References
-
- Affymetrix. 2001. GeneChip arrays for gene expression analysis. Technical note. Affymetrix, Santa Clara, CA.
-
- Affymetrix. 2001. GeneChip arrays provide optimal sensitivity and specificity for microarray expression analysis. Technical note. Affymetrix, Santa Clara, CA.
-
- Bibiloni, R., M. Mangold, K. L. Madsen, R. N. Fedorak, and G. W. Tannock. 2006. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn's disease and ulcerative colitis patients. J. Med. Microbiol. 55:1141-1149. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

