AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading
- PMID: 19499576
- PMCID: PMC3041641
- DOI: 10.1002/jcc.21334
AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading
Abstract
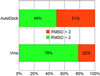
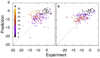
AutoDock Vina, a new program for molecular docking and virtual screening, is presented. AutoDock Vina achieves an approximately two orders of magnitude speed-up compared with the molecular docking software previously developed in our lab (AutoDock 4), while also significantly improving the accuracy of the binding mode predictions, judging by our tests on the training set used in AutoDock 4 development. Further speed-up is achieved from parallelism, by using multithreading on multicore machines. AutoDock Vina automatically calculates the grid maps and clusters the results in a way transparent to the user.
Copyright 2009 Wiley Periodicals, Inc.
Figures

Similar articles
-
QuickVina: accelerating AutoDock Vina using gradient-based heuristics for global optimization.IEEE/ACM Trans Comput Biol Bioinform. 2012 Sep-Oct;9(5):1266-72. doi: 10.1109/TCBB.2012.82. IEEE/ACM Trans Comput Biol Bioinform. 2012. PMID: 22641710
-
The Performance of Several Docking Programs at Reproducing Protein-Macrolide-Like Crystal Structures.Molecules. 2017 Jan 17;22(1):136. doi: 10.3390/molecules22010136. Molecules. 2017. PMID: 28106755 Free PMC article.
-
The scoring bias in reverse docking and the score normalization strategy to improve success rate of _target fishing.PLoS One. 2017 Feb 14;12(2):e0171433. doi: 10.1371/journal.pone.0171433. eCollection 2017. PLoS One. 2017. PMID: 28196116 Free PMC article.
-
Vinardo: A Scoring Function Based on Autodock Vina Improves Scoring, Docking, and Virtual Screening.PLoS One. 2016 May 12;11(5):e0155183. doi: 10.1371/journal.pone.0155183. eCollection 2016. PLoS One. 2016. PMID: 27171006 Free PMC article.
-
Survey of public domain software for docking simulations and virtual screening.Hum Genomics. 2011 Jul;5(5):497-505. doi: 10.1186/1479-7364-5-5-497. Hum Genomics. 2011. PMID: 21807604 Free PMC article. Review.
Cited by
-
Resveratrol Effect on α-Lactalbumin Thermal Stability.Biomedicines. 2024 Sep 25;12(10):2176. doi: 10.3390/biomedicines12102176. Biomedicines. 2024. PMID: 39457489 Free PMC article.
-
Isatin Bis-Imidathiazole Hybrids Identified as FtsZ Inhibitors with On-_target Activity Against Staphylococcus aureus.Antibiotics (Basel). 2024 Oct 19;13(10):992. doi: 10.3390/antibiotics13100992. Antibiotics (Basel). 2024. PMID: 39452258 Free PMC article.
-
In Silico Identification of Potential Inhibitors of SARS-CoV-2 Main Protease (Mpro).Pathogens. 2024 Oct 11;13(10):887. doi: 10.3390/pathogens13100887. Pathogens. 2024. PMID: 39452758 Free PMC article.
-
Hyrtios sp.-associated Cladosporium sp. UR3 as a potential source of antiproliferative metabolites.BMC Microbiol. 2024 Nov 1;24(1):445. doi: 10.1186/s12866-024-03560-6. BMC Microbiol. 2024. PMID: 39487417 Free PMC article.
-
Carvacrol potentiates immunity and sorafenib anti-cancer efficacy by _targeting HIF-1α/STAT3/ FGL1 pathway: in silico and in vivo study.Naunyn Schmiedebergs Arch Pharmacol. 2024 Oct 28. doi: 10.1007/s00210-024-03530-9. Online ahead of print. Naunyn Schmiedebergs Arch Pharmacol. 2024. PMID: 39466438
References
-
- Sousa SF, Fernandes PA, Ramos MJ. Protein-ligand docking: Current status and future challenges. Proteins-Structure Function and Bioinformatics. 2006 Oct 1;vol. 65:15–26. - PubMed
-
- Chang MW, Lindstrom W, Olson AJ, Belew RK. Analysis of HIV wild-type and mutant structures via in silico docking against diverse ligand libraries. Journal of Chemical Information and Modeling. 2007 May–Jun;vol. 47:1258–1262. - PubMed
-
- Gilson MK, Zhou H-X. Calculation of protein-ligand binding affinities. Annual Review of Biophysics and Biomolecular Structure. 2007;vol. 36:21–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

