Enhanced digestion efficiency, peptide ionization efficiency, and sequence resolution for protein hydrogen/deuterium exchange monitored by Fourier transform ion cyclotron resonance mass spectrometry
- PMID: 19551977
- PMCID: PMC2784605
- DOI: 10.1021/ac801417d
Enhanced digestion efficiency, peptide ionization efficiency, and sequence resolution for protein hydrogen/deuterium exchange monitored by Fourier transform ion cyclotron resonance mass spectrometry
Abstract
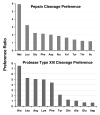
Solution-phase hydrogen/deuterium exchange (HDX) monitored by high-resolution Fourier transform ion cyclotron resonance (FTICR) mass spectrometry offers a rapid method to study protein conformations and protein-protein interactions. Pepsin is usually used to digest proteins in HDX and is known for lack of cleavage specificity. To improve digestion efficiency and specificity, we have optimized digestion conditions and cleavage preferences for pepsin and protease type XIII from Aspergillus saitoi. A dilution series of the proteases was used to determine the digestion efficiency for several test proteins. Protease type XIII prefers to cleave on the C-terminal end of basic amino acids and produced the highest number of fragments and the best sequence coverage compared to pepsin or protease type XVIII from Rhizhopus. Furthermore, protease type XIII exhibited much less self-digestion than pepsin and thus is superior for HDX experiments. Many highly overlapped segments from protease type XIII and pepsin digestion, combined with high-resolution FTICR mass spectrometry, provide high sequence resolution (to as few as one or two amino acids) for the assignment of amide hydrogen exchange rate. Our H/D exchange results correlate well with the secondary and tertiary structure of myoglobin. Such assignments of highly overlapped fragments promise to greatly enhance the accuracy and sequence resolution for determining conformational differences resulting from ligand binding or protein-protein interactions.
Figures




Similar articles
-
Local dynamics measured by hydrogen/deuterium exchange and mass spectrometry of creatine kinase digested by two proteases.Biochimie. 2005 Dec;87(12):1101-10. doi: 10.1016/j.biochi.2005.05.012. Epub 2005 Jun 29. Biochimie. 2005. PMID: 16023284
-
High-Resolution HDX-MS of Cytochrome c Using Pepsin/Fungal Protease Type XIII Mixed Bed Column.J Am Soc Mass Spectrom. 2019 Feb;30(2):227-234. doi: 10.1007/s13361-018-2087-7. Epub 2018 Oct 29. J Am Soc Mass Spectrom. 2019. PMID: 30374663
-
Use of different proteases working in acidic conditions to improve sequence coverage and resolution in hydrogen/deuterium exchange of large proteins.Rapid Commun Mass Spectrom. 2003;17(21):2387-93. doi: 10.1002/rcm.1207. Rapid Commun Mass Spectrom. 2003. PMID: 14587084
-
Measuring the hydrogen/deuterium exchange of proteins at high spatial resolution by mass spectrometry: overcoming gas-phase hydrogen/deuterium scrambling.Acc Chem Res. 2014 Oct 21;47(10):3018-27. doi: 10.1021/ar500194w. Epub 2014 Aug 29. Acc Chem Res. 2014. PMID: 25171396 Review.
-
Characterizing rapid, activity-linked conformational transitions in proteins via sub-second hydrogen deuterium exchange mass spectrometry.FEBS J. 2013 Nov;280(22):5616-25. doi: 10.1111/febs.12332. Epub 2013 Jun 11. FEBS J. 2013. PMID: 23663649 Review.
Cited by
-
Improved sequence resolution by global analysis of overlapped peptides in hydrogen/deuterium exchange mass spectrometry.J Am Soc Mass Spectrom. 2012 Jul;23(7):1202-8. doi: 10.1007/s13361-012-0373-3. Epub 2012 Apr 24. J Am Soc Mass Spectrom. 2012. PMID: 22528203
-
Uncovering of a short internal peptide activates a tRNA synthetase procytokine.J Biol Chem. 2012 Jun 8;287(24):20504-8. doi: 10.1074/jbc.C112.369439. Epub 2012 May 1. J Biol Chem. 2012. PMID: 22549774 Free PMC article.
-
Chasing Tails: Cathepsin-L Improves Structural Analysis of Histones by HX-MS.Mol Cell Proteomics. 2019 Oct;18(10):2089-2098. doi: 10.1074/mcp.RA119.001325. Epub 2019 Aug 13. Mol Cell Proteomics. 2019. PMID: 31409669 Free PMC article.
-
Orthogonal Mass Spectrometry-Based Footprinting for Epitope Mapping and Structural Characterization: The IL-6 Receptor upon Binding of Protein Therapeutics.Anal Chem. 2017 Jul 18;89(14):7742-7749. doi: 10.1021/acs.analchem.7b01748. Epub 2017 Jul 6. Anal Chem. 2017. PMID: 28621526 Free PMC article.
-
Determination of Equine Cytochrome c Backbone Amide Hydrogen/Deuterium Exchange Rates by Mass Spectrometry Using a Wider Time Window and Isotope Envelope.J Am Soc Mass Spectrom. 2017 Mar;28(3):486-497. doi: 10.1007/s13361-016-1571-1. Epub 2017 Jan 20. J Am Soc Mass Spectrom. 2017. PMID: 28108962
References
-
- Smith DL, Deng Y, Zhang Z. J Mass Spectrom. 1997;32:135–146. - PubMed
-
- Lanman JK, Lam TT, Barnes S, Sakalian M, Emmett MR, Marshall AG, Prevelige PE. J Mol Biol. 2003;325:759–772. - PubMed
-
- Lam TT, Lanman JK, Emmett MR, Hendrickson CL, Marshall AG, Prevelige PE. J Chromatogr A. 2002;982:85–95. - PubMed
-
- Engen JR, Smith DL. Anal Chem. 2001;73:256A–265A. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

