Atg32 is a mitochondrial protein that confers selectivity during mitophagy
- PMID: 19619495
- PMCID: PMC2746076
- DOI: 10.1016/j.devcel.2009.06.014
Atg32 is a mitochondrial protein that confers selectivity during mitophagy
Abstract
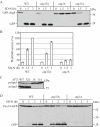
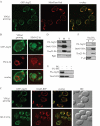
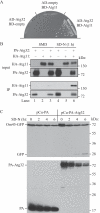
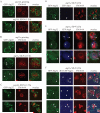
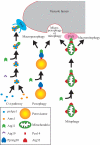
Mitochondrial quality control is important in maintaining proper cellular homeostasis. Although selective mitochondrial degradation by autophagy (mitophagy) is suggested to have an important role in quality control, and though there is evidence for a direct relation between mitophagy and neurodegenerative diseases, the molecular mechanism of mitophagy is poorly understood. Using a screen for mitophagy-deficient mutants, we found that YIL146C/ECM37 is essential for mitophagy. This gene is not required for other types of selective autophagy or for nonspecific macroautophagy. We designated this autophagy-related (ATG) gene as ATG32. The Atg32 protein localizes on mitochondria. Following the induction of mitophagy, Atg32 binds Atg11, an adaptor protein for selective types of autophagy, and is then recruited to and imported into the vacuole along with mitochondria. Therefore, Atg32 confers selectivity for mitochondrial sequestration as a cargo and is necessary for recruitment of this organelle by the autophagy machinery for mitophagy.
Figures

Comment in
-
A receptor for eating mitochondria.Dev Cell. 2009 Jul;17(1):1-2. doi: 10.1016/j.devcel.2009.07.003. Dev Cell. 2009. PMID: 19619484
Similar articles
-
Autophagy-related protein 32 acts as autophagic degron and directly initiates mitophagy.J Biol Chem. 2012 Mar 23;287(13):10631-10638. doi: 10.1074/jbc.M111.299917. Epub 2012 Feb 3. J Biol Chem. 2012. PMID: 22308029 Free PMC article.
-
Casein kinase 2 is essential for mitophagy.EMBO Rep. 2013 Sep;14(9):788-94. doi: 10.1038/embor.2013.114. Epub 2013 Jul 30. EMBO Rep. 2013. PMID: 23897086 Free PMC article.
-
Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy.Dev Cell. 2009 Jul;17(1):87-97. doi: 10.1016/j.devcel.2009.06.013. Dev Cell. 2009. PMID: 19619494
-
BCL2L13 is a mammalian homolog of the yeast mitophagy receptor Atg32.Autophagy. 2015;11(10):1932-3. doi: 10.1080/15548627.2015.1084459. Autophagy. 2015. PMID: 26506896 Free PMC article. Review.
-
Regulatory mechanisms of mitophagy in yeast.Biochim Biophys Acta Gen Subj. 2021 May;1865(5):129858. doi: 10.1016/j.bbagen.2021.129858. Epub 2021 Feb 3. Biochim Biophys Acta Gen Subj. 2021. PMID: 33545228 Review.
Cited by
-
Structural insights into the recognition of phosphorylated FUNDC1 by LC3B in mitophagy.Protein Cell. 2017 Jan;8(1):25-38. doi: 10.1007/s13238-016-0328-8. Epub 2016 Oct 18. Protein Cell. 2017. PMID: 27757847 Free PMC article.
-
The role of ALFY in selective autophagy.Cell Death Differ. 2013 Jan;20(1):12-20. doi: 10.1038/cdd.2012.66. Epub 2012 Jun 1. Cell Death Differ. 2013. PMID: 22653340 Free PMC article. Review.
-
Peroxisomal Pex3 activates selective autophagy of peroxisomes via interaction with the pexophagy receptor Atg30.J Biol Chem. 2015 Mar 27;290(13):8623-31. doi: 10.1074/jbc.M114.619338. Epub 2015 Feb 18. J Biol Chem. 2015. PMID: 25694426 Free PMC article.
-
Selective sorting and destruction of mitochondrial membrane proteins in aged yeast.Elife. 2016 Apr 20;5:e13943. doi: 10.7554/eLife.13943. Elife. 2016. PMID: 27097106 Free PMC article.
-
Phosphorylation of mitochondrial matrix proteins regulates their selective mitophagic degradation.Proc Natl Acad Sci U S A. 2019 Oct 8;116(41):20517-20527. doi: 10.1073/pnas.1901759116. Epub 2019 Sep 23. Proc Natl Acad Sci U S A. 2019. PMID: 31548421 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

