The role of MITF phosphorylation sites during coat color and eye development in mice analyzed by bacterial artificial chromosome transgene rescue
- PMID: 19635938
- PMCID: PMC2766318
- DOI: 10.1534/genetics.109.103945
The role of MITF phosphorylation sites during coat color and eye development in mice analyzed by bacterial artificial chromosome transgene rescue
Abstract
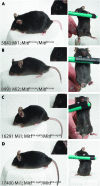
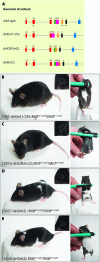
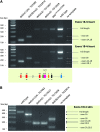
The microphthalmia-associated transcription factor (Mitf) has emerged as an important model for gene regulation in eukaryotic organisms. In vertebrates, it regulates the development of several cell types including melanocytes and has also been shown to play an important role in melanoma. In vitro, the activity of MITF is regulated by multiple signaling pathways, including the KITL/KIT/B-Raf pathway, which results in phosphorylation of MITF on serine residues 73 and 409. However, the precise role of signaling to MITF in vivo remains largely unknown. Here, we use a BAC transgene rescue approach to introduce specific mutations in MITF to study the importance of specific phospho-acceptor sites and protein domains. We show that mice that carry a BAC transgene where single-amino-acid substitutions have been made in the Mitf gene rescue the phenotype of the loss-of-function mutations in Mitf. This may indicate that signaling from KIT to MITF affects other phospho-acceptor sites in MITF or that alternative sites can be phosphorylated when Ser73 and Ser409 have been mutated. Our results have implications for understanding signaling to transcription factors. Furthermore, as MITF and signaling mechanisms have been shown to play an important role in melanomas, our findings may lead to novel insights into this resilient disease.
Figures




Similar articles
-
In vivo role of alternative splicing and serine phosphorylation of the microphthalmia-associated transcription factor.Genetics. 2012 May;191(1):133-44. doi: 10.1534/genetics.111.135996. Epub 2012 Feb 23. Genetics. 2012. PMID: 22367038 Free PMC article.
-
MicroRNA-25 functions in regulation of pigmentation by _targeting the transcription factor MITF in Alpaca (Lama pacos) skin melanocytes.Domest Anim Endocrinol. 2010 Apr;38(3):200-9. doi: 10.1016/j.domaniend.2009.10.004. Epub 2009 Nov 25. Domest Anim Endocrinol. 2010. PMID: 20036482
-
Melanocyte-lineage expression of Cre recombinase using Mitf regulatory elements.Pigment Cell Melanoma Res. 2008 Feb;21(1):63-9. doi: 10.1111/j.1755-148X.2007.00425.x. Pigment Cell Melanoma Res. 2008. PMID: 18353144
-
The Microphthalmia-Associated Transcription Factor (MITF) and Its Role in the Structure and Function of the Eye.Genes (Basel). 2024 Sep 27;15(10):1258. doi: 10.3390/genes15101258. Genes (Basel). 2024. PMID: 39457382 Free PMC article. Review.
-
MITF: a stream flowing for pigment cells.Pigment Cell Res. 2000 Aug;13(4):230-40. doi: 10.1034/j.1600-0749.2000.130404.x. Pigment Cell Res. 2000. PMID: 10952390 Review.
Cited by
-
RNAi-mediated SLC7A11 knockdown inhibits melanogenesis-related genes expression in rabbit skin fibroblasts.J Genet. 2018 Jun;97(2):463-468. J Genet. 2018. PMID: 29932066
-
Coat color determination by miR-137 mediated down-regulation of microphthalmia-associated transcription factor in a mouse model.RNA. 2012 Sep;18(9):1679-86. doi: 10.1261/rna.033977.112. Epub 2012 Jul 30. RNA. 2012. PMID: 22847819 Free PMC article.
-
MITF-the first 25 years.Genes Dev. 2019 Aug 1;33(15-16):983-1007. doi: 10.1101/gad.324657.119. Epub 2019 May 23. Genes Dev. 2019. PMID: 31123060 Free PMC article. Review.
-
Variation in pigmentation gene expression is associated with distinct aposematic color morphs in the poison frog Dendrobates auratus.BMC Evol Biol. 2019 Apr 18;19(1):85. doi: 10.1186/s12862-019-1410-7. BMC Evol Biol. 2019. PMID: 30995908 Free PMC article.
-
Characterization of POU2F1 Gene and Its Potential Impact on the Expression of Genes Involved in Fur Color Formation in Rex Rabbit.Genes (Basel). 2020 May 20;11(5):575. doi: 10.3390/genes11050575. Genes (Basel). 2020. PMID: 32443864 Free PMC article.
References
-
- Bharti, K., J. Debbache, J. Wang and H. Arnheiter, 2009. The basic helix-loop-helix leucine-zipper gene Mitf: analysis of alternative promoter choice and splicing in Methods in Molecular Biology: Transcription Factors, edited by Paul Higgins. Humana Press, Totowa, NJ (in press). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

