Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques
- PMID: 19743430
- PMCID: PMC2892724
- DOI: 10.1002/pmic.200900398
Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques
Abstract
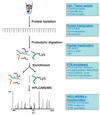
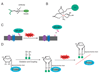
More than 300 different types of protein post-translational modifications (PTMs) have been described, many of which are known to have pivotal roles in cellular physiology and disease. Nevertheless, only a handful of PTMs have been extensively investigated at the proteome level. Knowledge of protein substrates and their PTM sites is key to dissection of PTM-mediated cellular processes. The past several years have seen a tremendous progress in developing MS-based proteomics technologies for global PTM analysis, including numerous studies of yeast and other microbes. Modification-specific enrichment techniques combined with advanced MS/MS methods and computational data analysis have revealed a surprisingly large extent of PTMs in proteins, including multi-site, cooperative modifications in individual proteins. We review some of the current strategies employed for enrichment and detection of PTMs in modification-specific proteomics.
Conflict of interest statement
The authors have declared no conflict of interest.
Figures
Similar articles
-
Identification, Quantification, and Site Localization of Protein Posttranslational Modifications via Mass Spectrometry-Based Proteomics.Adv Exp Med Biol. 2016;919:345-382. doi: 10.1007/978-3-319-41448-5_17. Adv Exp Med Biol. 2016. PMID: 27975226 Review.
-
The Methods Employed in Mass Spectrometric Analysis of Posttranslational Modifications (PTMs) and Protein-Protein Interactions (PPIs).Adv Exp Med Biol. 2019;1140:169-198. doi: 10.1007/978-3-030-15950-4_10. Adv Exp Med Biol. 2019. PMID: 31347048 Free PMC article. Review.
-
Advances in enrichment methods for mass spectrometry-based proteomics analysis of post-translational modifications.J Chromatogr A. 2022 Aug 16;1678:463352. doi: 10.1016/j.chroma.2022.463352. Epub 2022 Jul 19. J Chromatogr A. 2022. PMID: 35896048 Review.
-
Enrichment and separation techniques for large-scale proteomics analysis of the protein post-translational modifications.J Chromatogr A. 2014 Dec 12;1372C:1-17. doi: 10.1016/j.chroma.2014.10.107. Epub 2014 Nov 6. J Chromatogr A. 2014. PMID: 25465002 Review.
-
Substrate and Functional Diversity of Protein Lysine Post-translational Modifications.Genomics Proteomics Bioinformatics. 2024 May 9;22(1):qzae019. doi: 10.1093/gpbjnl/qzae019. Genomics Proteomics Bioinformatics. 2024. PMID: 38862432 Review.
Cited by
-
Advances in neuroproteomics for neurotrauma: unraveling insights for personalized medicine and future prospects.Front Neurol. 2023 Nov 22;14:1288740. doi: 10.3389/fneur.2023.1288740. eCollection 2023. Front Neurol. 2023. PMID: 38073638 Free PMC article. Review.
-
Dimer interface of bovine cytochrome c oxidase is influenced by local posttranslational modifications and lipid binding.Proc Natl Acad Sci U S A. 2016 Jul 19;113(29):8230-5. doi: 10.1073/pnas.1600354113. Epub 2016 Jun 30. Proc Natl Acad Sci U S A. 2016. PMID: 27364008 Free PMC article.
-
Statistical Analysis of Post-Translational Modifications Quantified by Label-Free Proteomics Across Multiple Biological Conditions with R: Illustration from SARS-CoV-2 Infected Cells.Methods Mol Biol. 2023;2426:267-302. doi: 10.1007/978-1-0716-1967-4_12. Methods Mol Biol. 2023. PMID: 36308693
-
Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study.J Biol Chem. 2010 Apr 9;285(15):11051-5. doi: 10.1074/jbc.R109.097600. Epub 2010 Feb 10. J Biol Chem. 2010. PMID: 20147750 Free PMC article. Review.
-
Mass spectrometry-based phosphoproteomics reveals multisite phosphorylation on mammalian brain voltage-gated sodium and potassium channels.Semin Cell Dev Biol. 2011 Apr;22(2):153-9. doi: 10.1016/j.semcdb.2010.09.009. Epub 2010 Oct 12. Semin Cell Dev Biol. 2011. PMID: 20932926 Free PMC article. Review.
References
-
- Gu W, Shi XL, Roeder RG. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. - PubMed
-
- Kim SC, Sprung R, Chen Y, Xu Y, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. - PubMed
-
- Zhu H, Klemic JF, Chang S, Bertone P, et al. Analysis of yeast protein kinases using protein chips. Nat. Genet. 2000;26:283–289. - PubMed
-
- Reimer U, Reineke U, Schneider-Mergener J. Peptide arrays: from macro to micro. Curr. Opin. Biotechnol. 2002;13:315–320. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

