JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin
- PMID: 19783980
- PMCID: PMC3785147
- DOI: 10.1038/nature08448
JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin
Abstract
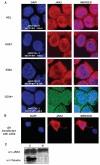
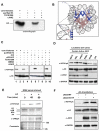
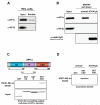
Activation of Janus kinase 2 (JAK2) by chromosomal translocations or point mutations is a frequent event in haematological malignancies. JAK2 is a non-receptor tyrosine kinase that regulates several cellular processes by inducing cytoplasmic signalling cascades. Here we show that human JAK2 is present in the nucleus of haematopoietic cells and directly phosphorylates Tyr 41 (Y41) on histone H3. Heterochromatin protein 1alpha (HP1alpha), but not HP1beta, specifically binds to this region of H3 through its chromo-shadow domain. Phosphorylation of H3Y41 by JAK2 prevents this binding. Inhibition of JAK2 activity in human leukaemic cells decreases both the expression of the haematopoietic oncogene lmo2 and the phosphorylation of H3Y41 at its promoter, while simultaneously increasing the binding of HP1alpha at the same site. Tauhese results identify a previously unrecognized nuclear role for JAK2 in the phosphorylation of H3Y41 and reveal a direct mechanistic link between two genes, jak2 and lmo2, involved in normal haematopoiesis and leukaemia.
Figures





Similar articles
-
Janus kinase 2: an epigenetic 'writer' that activates leukemogenic genes.J Mol Cell Biol. 2010 Oct;2(5):231-3. doi: 10.1093/jmcb/mjp054. Epub 2010 Jan 5. J Mol Cell Biol. 2010. PMID: 20051435
-
LIF-independent JAK signalling to chromatin in embryonic stem cells uncovered from an adult stem cell disease.Nat Cell Biol. 2011 Jan;13(1):13-21. doi: 10.1038/ncb2135. Epub 2010 Dec 12. Nat Cell Biol. 2011. PMID: 21151131 Free PMC article.
-
Nuclear localization of lymphocyte-specific protein tyrosine kinase (Lck) and its role in regulating LIM domain only 2 (Lmo2) gene.Biochem Biophys Res Commun. 2012 Jan 20;417(3):1058-62. doi: 10.1016/j.bbrc.2011.12.095. Epub 2011 Dec 27. Biochem Biophys Res Commun. 2012. PMID: 22222369 Free PMC article.
-
JAK2 gets histone H3 rolling.Cancer Cell. 2009 Nov 6;16(5):365-6. doi: 10.1016/j.ccr.2009.10.009. Cancer Cell. 2009. PMID: 19878867 Review.
-
Beyond the histone tale: HP1α deregulation in breast cancer epigenetics.Cancer Biol Ther. 2015;16(2):189-200. doi: 10.1080/15384047.2014.1001277. Cancer Biol Ther. 2015. PMID: 25588111 Free PMC article. Review.
Cited by
-
Comprehensive review of JAK inhibitors in myeloproliferative neoplasms.Ther Adv Hematol. 2013 Feb;4(1):15-35. doi: 10.1177/2040620712461047. Ther Adv Hematol. 2013. PMID: 23610611 Free PMC article.
-
Mechanisms of Jak/STAT signaling in immunity and disease.J Immunol. 2015 Jan 1;194(1):21-7. doi: 10.4049/jimmunol.1401867. J Immunol. 2015. PMID: 25527793 Free PMC article. Review.
-
High-order chromatin structure and the epigenome in SAHFs.Nucleus. 2013 Jan-Feb;4(1):23-8. doi: 10.4161/nucl.23189. Epub 2012 Dec 11. Nucleus. 2013. PMID: 23232545 Free PMC article.
-
Histone H3 phosphorylation - a versatile chromatin modification for different occasions.Biochimie. 2012 Nov;94(11):2193-201. doi: 10.1016/j.biochi.2012.04.018. Epub 2012 Apr 28. Biochimie. 2012. PMID: 22564826 Free PMC article. Review.
-
JAK2 in Myeloproliferative Neoplasms: Still a Protagonist.Pharmaceuticals (Basel). 2022 Jan 28;15(2):160. doi: 10.3390/ph15020160. Pharmaceuticals (Basel). 2022. PMID: 35215273 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

