Antibiotic resistance genes in the vaginal microbiota of primates not normally exposed to antibiotics
- PMID: 19857138
- PMCID: PMC3145952
- DOI: 10.1089/mdr.2009.0052
Antibiotic resistance genes in the vaginal microbiota of primates not normally exposed to antibiotics
Abstract
Previous studies of resistance gene ecology have focused primarily on populations such as hospital patients and farm animals that are regularly exposed to antibiotics. Also, these studies have tended to focus on numerically minor populations such as enterics or enterococci. We report here a cultivation-independent approach that allowed us to assess the presence of antibiotic resistance genes in the numerically predominant populations of the vaginal microbiota of two populations of primates that are seldom or never exposed to antibiotics: baboons and mangabeys. Most of these animals were part of a captive colony in Texas that is used for scientific studies of female physiology and physical anthropology topics. Samples from some wild baboons were also tested. Vaginal swab samples, obtained in connection with a study designed to define the normal microbiota of the female vaginal canal, were tested for the presence of two types of antibiotic resistance genes: tetracycline resistance (tet) genes and erythromycin resistance (erm) genes. These genes are frequently found in human isolates of the two types of bacteria that were a substantial part of the normal microbiota of primates (Firmicutes and Bacteroidetes). Since cultivation was not feasible, polymerase chain reaction and DNA sequencing were used to detect and characterize these resistance genes. The tet(M) and tet(W) genes were found most commonly, and the tet(Q) gene was found in over a third of the samples from baboons. The ermB and ermF genes were found only in a minority of the samples. The ermG gene was not found in any of the specimens tested. Polymerase chain reaction analysis showed that at least some tet(M) and tet(Q) genes were genetically linked to DNA from known conjugative transposons (CTns), Tn916 and CTnDOT. Our results raise questions about the extent to which extensive exposure to antibiotics is the only pressure necessary to maintain resistance genes in natural settings.
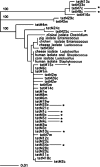
Figures
Similar articles
-
Distribution of erm(F) and tet(Q) genes in 4 oral bacterial species and genotypic variation between resistant and susceptible isolates.J Clin Periodontol. 2002 Feb;29(2):152-8. doi: 10.1034/j.1600-051x.2002.290210.x. J Clin Periodontol. 2002. PMID: 11895543
-
Presence of the tet(O) gene in erythromycin- and tetracycline-resistant strains of Streptococcus pyogenes and linkage with either the mef(A) or the erm(A) gene.Antimicrob Agents Chemother. 2003 Sep;47(9):2844-9. doi: 10.1128/AAC.47.9.2844-2849.2003. Antimicrob Agents Chemother. 2003. PMID: 12936983 Free PMC article.
-
Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon.Appl Environ Microbiol. 2001 Feb;67(2):561-8. doi: 10.1128/AEM.67.2.561-568.2001. Appl Environ Microbiol. 2001. PMID: 11157217 Free PMC article.
-
Detection of a genetic linkage between genes coding for resistance to tetracycline and erythromycin in Clostridium difficile.Microb Drug Resist. 2007 Summer;13(2):90-5. doi: 10.1089/mdr.2007.723. Microb Drug Resist. 2007. PMID: 17650959
-
Prevalence of antibiotic resistance genes in the oral cavity and mobile genetic elements that disseminate antimicrobial resistance: A systematic review.Mol Oral Microbiol. 2022 Aug;37(4):133-153. doi: 10.1111/omi.12375. Epub 2022 Jun 17. Mol Oral Microbiol. 2022. PMID: 35674142 Review.
Cited by
-
A case of Lactobacillus jensenii associated native valve endocarditis.IDCases. 2023 May 18;32:e01806. doi: 10.1016/j.idcr.2023.e01806. eCollection 2023. IDCases. 2023. PMID: 37250380 Free PMC article.
-
An Epidemiologic Study of Bacterial Culture and Antibiotic Susceptibility Analyses in Captive Macaques and Marmosets at the Wisconsin National Primate Research Center.J Am Assoc Lab Anim Sci. 2024 Apr 22;63(5):540-51. doi: 10.30802/AALAS-JAALAS-23-000079. Online ahead of print. J Am Assoc Lab Anim Sci. 2024. PMID: 38649259
-
Vaginal Dysbiosis from an Evolutionary Perspective.Sci Rep. 2016 May 26;6:26817. doi: 10.1038/srep26817. Sci Rep. 2016. PMID: 27226349 Free PMC article.
-
Functional metagenomics reveals previously unrecognized diversity of antibiotic resistance genes in gulls.Front Microbiol. 2011 Nov 29;2:238. doi: 10.3389/fmicb.2011.00238. eCollection 2011. Front Microbiol. 2011. PMID: 22347872 Free PMC article.
-
Distribution of ermB, ermF, tet(W), and tet(M) Resistance Genes in the Vaginal Ecosystem of Women during Pregnancy and Puerperium.Pathogens. 2021 Nov 26;10(12):1546. doi: 10.3390/pathogens10121546. Pathogens. 2021. PMID: 34959501 Free PMC article.
References
-
- Angulo F.J. Baker N.L. Olsen S.J. Anderson A. Barrett T.J. Antimicrobial use in agriculture: controlling the transfer of antimicrobial resistance to humans. Semin. Pediatr. Infect. Dis. 2004;15:78–85. - PubMed
-
- Celli J. Trieu-Cuot P. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol. Microbiol. 1998;28:103–117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

