Retinoic acid mediates long-paced oscillations in retinoid receptor activity: evidence for a potential role for RIP140
- PMID: 19862326
- PMCID: PMC2763268
- DOI: 10.1371/journal.pone.0007639
Retinoic acid mediates long-paced oscillations in retinoid receptor activity: evidence for a potential role for RIP140
Abstract
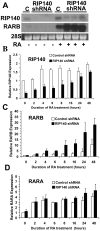
Background: Mechanisms that underlie oscillatory transcriptional activity of nuclear receptors (NRs) are incompletely understood. Evidence exists for rapid, cyclic recruitment of coregulatory complexes upon activation of nuclear receptors. RIP140 is a NR coregulator that represses the transactivation of agonist-bound nuclear receptors. Previously, we showed that RIP140 is inducible by all-trans retinoic acid (RA) and mediates limiting, negative-feedback regulation of retinoid signaling.
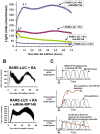
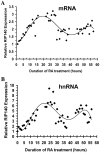
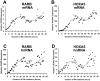
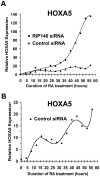
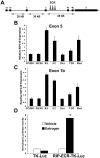
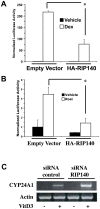
Methodology and findings: Here we report that in the continued presence of RA, long-paced oscillations of retinoic acid receptor (RAR) activity occur with a period ranging from 24 to 35 hours. Endogenous expression of RIP140 and other RA-_target genes also oscillate in the presence of RA. Cyclic retinoid receptor transactivation is ablated by constitutive overexpression of RIP140. Further, depletion of RIP140 disrupts cyclic expression of the RA _target gene HOXA5. Evidence is provided that RIP140 may limit RAR signaling in a selective, non-redundant manner in contrast to the classic NR coregulators NCoR1 and SRC1 that are not RA-inducible, do not cycle, and may be partially redundant in limiting RAR activity. Finally, evidence is provided that RIP140 can repress and be induced by other nuclear receptors in a manner that suggests potential participation in other NR oscillations.
Conclusions and significance: We provide evidence for novel, long-paced oscillatory retinoid receptor activity and hypothesize that this may be paced in part, by RIP140. Oscillatory NR activity may be involved in mediating hormone actions of physiological and pathological importance.
Conflict of interest statement
Figures

Similar articles
-
Negative feedback at the level of nuclear receptor coregulation. Self-limitation of retinoid signaling by RIP140.J Biol Chem. 2003 Nov 7;278(45):43889-92. doi: 10.1074/jbc.C300374200. Epub 2003 Sep 22. J Biol Chem. 2003. PMID: 14506269
-
Characterization of receptor-interacting protein 140 in retinoid receptor activities.J Biol Chem. 1999 Oct 29;274(44):31320-6. doi: 10.1074/jbc.274.44.31320. J Biol Chem. 1999. PMID: 10531331
-
Suppressive effect of receptor-interacting protein 140 on coregulator binding to retinoic acid receptor complexes, histone-modifying enzyme activity, and gene activation.J Biol Chem. 2004 Jan 2;279(1):319-25. doi: 10.1074/jbc.M307621200. Epub 2003 Oct 27. J Biol Chem. 2004. PMID: 14581481
-
Retinoids and receptor interacting protein 140 (RIP140) in gene regulation.Curr Med Chem. 2004 Jun;11(12):1527-32. doi: 10.2174/0929867043365017. Curr Med Chem. 2004. PMID: 15180561 Review.
-
Role of RIP140 in metabolic tissues: connections to disease.FEBS Lett. 2008 Jan 9;582(1):39-45. doi: 10.1016/j.febslet.2007.11.017. Epub 2007 Nov 20. FEBS Lett. 2008. PMID: 18023280 Review.
Cited by
-
Modulation of clock gene expression by the transcriptional coregulator receptor interacting protein 140 (RIP140).J Biol Rhythms. 2011 Jun;26(3):187-99. doi: 10.1177/0748730411401579. J Biol Rhythms. 2011. PMID: 21628546 Free PMC article.
-
Downregulation of RIP140 in hepatocellular carcinoma promoted the growth and migration of the cancer cells.Tumour Biol. 2015 Mar;36(3):2077-85. doi: 10.1007/s13277-014-2815-y. Epub 2014 Nov 13. Tumour Biol. 2015. PMID: 25391428
-
Headway and hurdles in the clinical development of dietary phytochemicals for cancer therapy and prevention: lessons learned from vitamin A derivatives.AAPS J. 2014 Mar;16(2):281-8. doi: 10.1208/s12248-014-9562-2. Epub 2014 Jan 16. AAPS J. 2014. PMID: 24431081 Free PMC article. Review.
-
Splicing-dependent RNA polymerase pausing in yeast.Mol Cell. 2010 Nov 24;40(4):582-93. doi: 10.1016/j.molcel.2010.11.005. Mol Cell. 2010. PMID: 21095588 Free PMC article.
-
Correlation between receptor-interacting protein 140 expression and directed differentiation of human embryonic stem cells into neural stem cells.Neural Regen Res. 2017 Jan;12(1):118-124. doi: 10.4103/1673-5374.198997. Neural Regen Res. 2017. PMID: 28250757 Free PMC article.
References
-
- Niles RM. Signaling pathways in retinoid chemoprevention and treatment of cancer. Mutat Res. 2004;555:81–96. - PubMed
-
- Freemantle SJ, Spinella MJ, Dmitrovsky E. Retinoids in cancer therapy and prevention: Promise meets resistance. Oncogene. 2003;22:7305–7315. - PubMed
-
- Buletic Z, Soprano KJ, Soprano DR. Retinoid _targets for the treatment of cancer. Crit Rev Eukaryot Gene Exp. 2006;16:193–210. - PubMed
-
- Perissi V, Rosenfeld MG. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol. 2005;7:542–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

