Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics
- PMID: 20062526
- PMCID: PMC2796172
- DOI: 10.1371/journal.ppat.1000719
Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics
Abstract
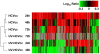
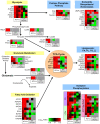
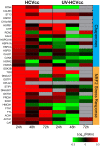
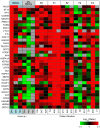
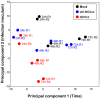
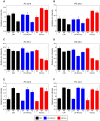
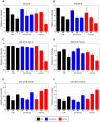
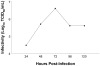
Proteomic and lipidomic profiling was performed over a time course of acute hepatitis C virus (HCV) infection in cultured Huh-7.5 cells to gain new insights into the intracellular processes influenced by this virus. Our proteomic data suggest that HCV induces early perturbations in glycolysis, the pentose phosphate pathway, and the citric acid cycle, which favor host biosynthetic activities supporting viral replication and propagation. This is followed by a compensatory shift in metabolism aimed at maintaining energy homeostasis and cell viability during elevated viral replication and increasing cellular stress. Complementary lipidomic analyses identified numerous temporal perturbations in select lipid species (e.g. phospholipids and sphingomyelins) predicted to play important roles in viral replication and downstream assembly and secretion events. The elevation of lipotoxic ceramide species suggests a potential link between HCV-associated biochemical alterations and the direct cytopathic effect observed in this in vitro system. Using innovative computational modeling approaches, we further identified mitochondrial fatty acid oxidation enzymes, which are comparably regulated during in vitro infection and in patients with histological evidence of fibrosis, as possible _targets through which HCV regulates temporal alterations in cellular metabolic homeostasis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Pyruvate dehydrogenase kinase regulates hepatitis C virus replication.Sci Rep. 2016 Jul 29;6:30846. doi: 10.1038/srep30846. Sci Rep. 2016. PMID: 27471054 Free PMC article.
-
Oxidative Stress Attenuates Lipid Synthesis and Increases Mitochondrial Fatty Acid Oxidation in Hepatoma Cells Infected with Hepatitis C Virus.J Biol Chem. 2016 Jan 22;291(4):1974-1990. doi: 10.1074/jbc.M115.674861. Epub 2015 Dec 1. J Biol Chem. 2016. PMID: 26627833 Free PMC article.
-
N-Myc Downstream-Regulated Gene 1 Restricts Hepatitis C Virus Propagation by Regulating Lipid Droplet Biogenesis and Viral Assembly.J Virol. 2018 Jan 2;92(2):e01166-17. doi: 10.1128/JVI.01166-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29118118 Free PMC article.
-
Hepatitis C virus hijacks host lipid metabolism.Trends Endocrinol Metab. 2010 Jan;21(1):33-40. doi: 10.1016/j.tem.2009.07.005. Epub 2009 Oct 23. Trends Endocrinol Metab. 2010. PMID: 19854061 Free PMC article. Review.
-
Whole Lotta Lipids-from HCV RNA Replication to the Mature Viral Particle.Int J Mol Sci. 2020 Apr 21;21(8):2888. doi: 10.3390/ijms21082888. Int J Mol Sci. 2020. PMID: 32326151 Free PMC article. Review.
Cited by
-
Mitochondria _targeted Viral Replication and Survival Strategies-Prospective on SARS-CoV-2.Front Pharmacol. 2020 Aug 28;11:578599. doi: 10.3389/fphar.2020.578599. eCollection 2020. Front Pharmacol. 2020. PMID: 32982760 Free PMC article. Review.
-
Systems approaches to influenza-virus host interactions and the pathogenesis of highly virulent and pandemic viruses.Semin Immunol. 2013 Oct 31;25(3):228-39. doi: 10.1016/j.smim.2012.11.001. Epub 2012 Dec 5. Semin Immunol. 2013. PMID: 23218769 Free PMC article. Review.
-
A systems biology starter kit for arenaviruses.Viruses. 2012 Dec;4(12):3625-46. doi: 10.3390/v4123625. Viruses. 2012. PMID: 23342371 Free PMC article. Review.
-
Reverse enGENEering of Regulatory Networks from Big Data: A Roadmap for Biologists.Bioinform Biol Insights. 2015 Apr 29;9:61-74. doi: 10.4137/BBI.S12467. eCollection 2015. Bioinform Biol Insights. 2015. PMID: 25983554 Free PMC article. Review.
-
Topological analysis of protein co-abundance networks identifies novel host _targets important for HCV infection and pathogenesis.BMC Syst Biol. 2012 Apr 30;6:28. doi: 10.1186/1752-0509-6-28. BMC Syst Biol. 2012. PMID: 22546282 Free PMC article.
References
-
- Alter MJ, Margolis HS, Krawczynski K, Judson FN, Mares A, et al. The natural history of community-acquired hepatitis C in the United States. The Sentinel Counties Chronic non-A, non-B Hepatitis Study Team. N Engl J Med. 1992;327:1899–1905. - PubMed
-
- He Y, Duan W, Tan SL. Emerging host cell _targets for hepatitis C therapy. Drug Discov Today. 2007;12:209–217. - PubMed
-
- Ye J. Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus. PLoS Pathog. 2007;3:e108. doi: 10.1371/journal.ppat.0030108. - DOI - PMC - PubMed
-
- Negro F, Sanyal AJ. Hepatitis C virus, steatosis and lipid abnormalities: clinical and pathogenic data. Liver Int. 2009;S2:26–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

