A Coronin7 homolog with functions in actin-driven processes
- PMID: 20071332
- PMCID: PMC2838343
- DOI: 10.1074/jbc.M109.083725
A Coronin7 homolog with functions in actin-driven processes
Abstract
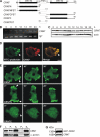
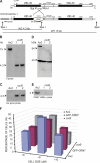
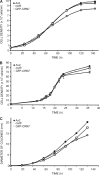
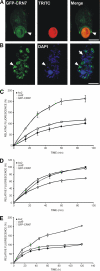
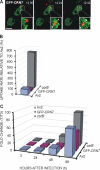
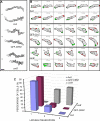
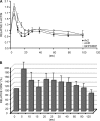
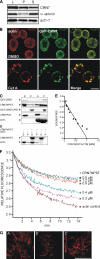
Dictyostelium discoideum Coronin7 (DdCRN7) together with human Coronin7 (CRN7) and Pod-1 of Drosophila melanogaster and Caenorhabditis elegans belong to the coronin family of WD-repeat domain-containing proteins. Coronin7 proteins are characterized by two WD-repeat domains that presumably fold into two beta-propeller structures. DdCRN7 shares highest homology with human CRN7, a protein with roles in membrane trafficking. DdCRN7 is present in the cytosol and accumulates in cell surface projections during movement and phago- and pinocytosis. Cells lacking CRN7 have altered chemotaxis and phagocytosis. Furthermore, loss of CRN7 affects the infection process by the pathogen Legionella pneumophila and allows a more efficient internalization of bacteria. To provide a mechanism for CNR7 action, we studied actin-related aspects. We could show that CRN7 binds directly to F-actin and protects actin filaments from depolymerization. CRN7 also associated with F-actin in vivo. It was present in the Triton X-100-insoluble cytoskeleton, colocalized with F-actin, and its distribution was sensitive to drugs affecting the actin cytoskeleton. We propose that the CRN7 role in chemotaxis and phagocytosis is through its effect on the actin cytoskeleton.
Figures
Similar articles
-
Redundant and unique roles of coronin proteins in Dictyostelium.Cell Mol Life Sci. 2011 Jan;68(2):303-13. doi: 10.1007/s00018-010-0455-y. Epub 2010 Jul 18. Cell Mol Life Sci. 2011. PMID: 20640912 Free PMC article.
-
Novel Coronin7 interactions with Cdc42 and N-WASP regulate actin organization and Golgi morphology.Sci Rep. 2016 May 4;6:25411. doi: 10.1038/srep25411. Sci Rep. 2016. PMID: 27143109 Free PMC article.
-
Coronin7 regulates WASP and SCAR through CRIB mediated interaction with Rac proteins.Sci Rep. 2015 Sep 28;5:14437. doi: 10.1038/srep14437. Sci Rep. 2015. PMID: 26411260 Free PMC article.
-
The Dictyostelium cytoskeleton.Experientia. 1995 Dec 18;51(12):1135-43. doi: 10.1007/BF01944731. Experientia. 1995. PMID: 8536801 Review.
-
Coronin proteins as multifunctional regulators of the cytoskeleton and membrane trafficking.Bioessays. 2005 Jun;27(6):625-32. doi: 10.1002/bies.20235. Bioessays. 2005. PMID: 15892111 Review.
Cited by
-
Actin filament debranching regulates cell polarity during cell migration and asymmetric cell division.Proc Natl Acad Sci U S A. 2021 Sep 14;118(37):e2100805118. doi: 10.1073/pnas.2100805118. Proc Natl Acad Sci U S A. 2021. PMID: 34507987 Free PMC article.
-
Unraveling the enigma: progress towards understanding the coronin family of actin regulators.Trends Cell Biol. 2011 Aug;21(8):481-8. doi: 10.1016/j.tcb.2011.04.004. Epub 2011 Jun 1. Trends Cell Biol. 2011. PMID: 21632254 Free PMC article. Review.
-
The professional phagocyte Dictyostelium discoideum as a model host for bacterial pathogens.Curr Drug _targets. 2011 Jun;12(7):942-54. doi: 10.2174/138945011795677782. Curr Drug _targets. 2011. PMID: 21366522 Free PMC article. Review.
-
Regulation of the Actin Cytoskeleton via Rho GTPase Signalling in Dictyostelium and Mammalian Cells: A Parallel Slalom.Cells. 2021 Jun 24;10(7):1592. doi: 10.3390/cells10071592. Cells. 2021. PMID: 34202767 Free PMC article. Review.
-
Cul3-KLHL20 ubiquitin ligase: physiological functions, stress responses, and disease implications.Cell Div. 2016 Apr 1;11:5. doi: 10.1186/s13008-016-0017-2. eCollection 2016. Cell Div. 2016. PMID: 27042198 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

