A mouse model of osteochondromagenesis from clonal inactivation of Ext1 in chondrocytes
- PMID: 20080592
- PMCID: PMC2836675
- DOI: 10.1073/pnas.0910875107
A mouse model of osteochondromagenesis from clonal inactivation of Ext1 in chondrocytes
Abstract
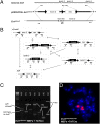
We report a mouse model of multiple osteochondromas (MO), an autosomal dominant disease in humans, also known as multiple hereditary exostoses (MHE or HME) and characterized by the formation of cartilage-capped osseous growths projecting from the metaphyses of endochondral bones. The pathogenesis of these osteochondromas has remained unclear. Mice heterozygous for Ext1 or Ext2, modeling the human genotypes that cause MO, occasionally develop solitary osteochondroma-like structures on ribs [Lin et al. (2000) Dev Biol 224(2):299-311; Stickens et al. (2005) Development 132(22):5055-5068]. Rather than model the germ-line genotype, we modeled the chimeric tissue genotype of somatic loss of heterozygosity (LOH), by conditionally inactivating Ext1 via head-to-head loxP sites and temporally controlled Cre-recombinase in chondrocytes. These mice faithfully recapitulate the human phenotype of multiple metaphyseal osteochondromas. We also confirm homozygous disruption of Ext1 in osteochondroma chondrocytes and their origin in proliferating physeal chondrocytes. These results explain prior modeling failures with the necessity for somatic LOH in a developmentally regulated cell type.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Comment in
-
EXTra hit for mouse osteochondroma.Proc Natl Acad Sci U S A. 2010 Feb 2;107(5):1813-4. doi: 10.1073/pnas.0914431107. Proc Natl Acad Sci U S A. 2010. PMID: 20133829 Free PMC article. No abstract available.
Similar articles
-
Toward an understanding of the short bone phenotype associated with multiple osteochondromas.J Orthop Res. 2013 Apr;31(4):651-7. doi: 10.1002/jor.22280. Epub 2012 Nov 28. J Orthop Res. 2013. PMID: 23192691 Free PMC article.
-
A mouse model of chondrocyte-specific somatic mutation reveals a role for Ext1 loss of heterozygosity in multiple hereditary exostoses.Proc Natl Acad Sci U S A. 2010 Jun 15;107(24):10932-7. doi: 10.1073/pnas.0914642107. Epub 2010 Jun 1. Proc Natl Acad Sci U S A. 2010. PMID: 20534475 Free PMC article.
-
Signaling systems affecting the severity of multiple osteochondromas.Bone. 2018 Jun;111:71-81. doi: 10.1016/j.bone.2018.03.010. Epub 2018 Mar 13. Bone. 2018. PMID: 29545125
-
The pathogenic roles of heparan sulfate deficiency in hereditary multiple exostoses.Matrix Biol. 2018 Oct;71-72:28-39. doi: 10.1016/j.matbio.2017.12.011. Epub 2017 Dec 24. Matrix Biol. 2018. PMID: 29277722 Free PMC article. Review.
-
Multiple osteochondromas.Orphanet J Rare Dis. 2008 Feb 13;3:3. doi: 10.1186/1750-1172-3-3. Orphanet J Rare Dis. 2008. PMID: 18271966 Free PMC article. Review.
Cited by
-
Inactivation of Fam20B in Joint Cartilage Leads to Chondrosarcoma and Postnatal Ossification Defects.Sci Rep. 2016 Jul 13;6:29814. doi: 10.1038/srep29814. Sci Rep. 2016. PMID: 27405802 Free PMC article.
-
Genetic and functional analyses detect an EXT1 splicing pathogenic variant in a Chinese hereditary multiple exostosis (HME) family.Mol Genet Genomic Med. 2022 Mar;10(3):e1878. doi: 10.1002/mgg3.1878. Epub 2022 Feb 1. Mol Genet Genomic Med. 2022. PMID: 35106951 Free PMC article.
-
Osteochondroma formation is independent of heparanase expression as revealed in a mouse model of hereditary multiple exostoses.J Orthop Res. 2022 Oct;40(10):2391-2401. doi: 10.1002/jor.25260. Epub 2022 Jan 22. J Orthop Res. 2022. PMID: 34996123 Free PMC article.
-
Heparan sulfate antagonism alters bone morphogenetic protein signaling and receptor dynamics, suggesting a mechanism in hereditary multiple exostoses.J Biol Chem. 2018 May 18;293(20):7703-7716. doi: 10.1074/jbc.RA117.000264. Epub 2018 Apr 5. J Biol Chem. 2018. PMID: 29622677 Free PMC article.
-
Toward an understanding of the short bone phenotype associated with multiple osteochondromas.J Orthop Res. 2013 Apr;31(4):651-7. doi: 10.1002/jor.22280. Epub 2012 Nov 28. J Orthop Res. 2013. PMID: 23192691 Free PMC article.
References
-
- Wicklund CL, Pauli RM, Johnston D, Hecht JT. Natural history study of hereditary multiple exostoses. Am J Med Genet. 1995;55:43–46. - PubMed
-
- Ahn J, et al. Cloning of the putative tumour suppressor gene for hereditary multiple exostoses (EXT1) Nat Genet. 1995;11:137–143. - PubMed
-
- Ligon AH, Potocki L, Shaffer LG, Stickens D, Evans GA. Gene for multiple exostoses (EXT2) maps to 11(p11.2p12) and is deleted in patients with a contiguous gene syndrome. Am J Med Genet. 1998;75:538–540. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

