Bone morphogenetic protein-7 inhibits proximal tubular epithelial cell Smad3 signaling via increased SnoN expression
- PMID: 20093492
- PMCID: PMC2832137
- DOI: 10.2353/ajpath.2010.090459
Bone morphogenetic protein-7 inhibits proximal tubular epithelial cell Smad3 signaling via increased SnoN expression
Abstract
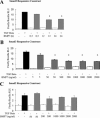
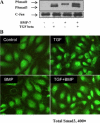
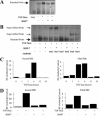
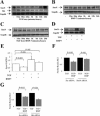
Bone morphogenetic protein-7 (BMP-7) improves outcome in animal models of fibrotic renal disease by opposing transforming growth factor beta1 (TGF-beta)-dependent fibrosis. However, the underlying mechanisms remain obscure. Here, we studied the effect of BMP-7 on response to TGF-beta in the proximal tubular cell line HK-2 (PTC). BMP-7 specifically limited Smad3 but not Smad2 signaling. BMP-7 did not inhibit Smad3 phosphorylation or nuclear accumulation, nor did BMP-7 alter phosphorylated Smad3 dephosphorylation or degradation. However, BMP-7 treatment reduced Smad3 DNA binding to a consensus Smad binding element probe, and chromatin immunoprecipitation showed reduced Smad3 binding to the plasminogen activator inhibitor-1 promoter in PTCs treated with BMP-7 and TGF-beta compared with TGF-beta alone. Degradation of the transcriptional repressor SnoN has recently been shown to be necessary for Smad3 (but not Smad2) signaling. SnoN expression was transiently lost in PTCs after TGF-beta stimulation, but BMP-7 prevented this. Furthermore, BMP-7 had no effect on Smad3 signaling after siRNA-mediated SnoN knockdown, whereas prevention of SnoN degradation with the proteasome inhibitor MG132 reproduced the inhibitory action of BMP-7 on Smad3 signaling. We conclude that BMP-7 prevents TGF-beta-mediated loss of the transcriptional repressor SnoN and hence specifically limits Smad3 DNA binding, altering the balance of transcriptional responses to TGF-beta in PTCs. These results provide an important mechanistic insight into a key regulator of TGF-beta signaling.
Figures



Similar articles
-
Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation.Mol Cell Biol. 2007 Sep;27(17):6068-83. doi: 10.1128/MCB.00664-07. Epub 2007 Jun 25. Mol Cell Biol. 2007. PMID: 17591695 Free PMC article.
-
A novel mechanism by which hepatocyte growth factor blocks tubular epithelial to mesenchymal transition.J Am Soc Nephrol. 2005 Jan;16(1):68-78. doi: 10.1681/ASN.2003090795. Epub 2004 Nov 10. J Am Soc Nephrol. 2005. PMID: 15537870
-
The downregulation of SnoN expression in human renal proximal tubule epithelial cells under high-glucose conditions is mediated by an increase in Smurf2 expression through TGF-β1 signaling.Int J Mol Med. 2016 Feb;37(2):415-22. doi: 10.3892/ijmm.2015.2448. Epub 2015 Dec 31. Int J Mol Med. 2016. PMID: 26743567
-
Role of the TGF-β/BMP-7/Smad pathways in renal diseases.Clin Sci (Lond). 2013 Feb;124(4):243-54. doi: 10.1042/CS20120252. Clin Sci (Lond). 2013. PMID: 23126427 Review.
-
Smad anchor for receptor activation (SARA) in TGF-beta signaling.Front Biosci (Elite Ed). 2010 Jun 1;2(3):857-60. doi: 10.2741/e147. Front Biosci (Elite Ed). 2010. PMID: 20515759 Review.
Cited by
-
TGF-β/Smad signaling in renal fibrosis.Front Physiol. 2015 Mar 19;6:82. doi: 10.3389/fphys.2015.00082. eCollection 2015. Front Physiol. 2015. PMID: 25852569 Free PMC article. Review.
-
Roles of the TGF-β⁻VEGF-C Pathway in Fibrosis-Related Lymphangiogenesis.Int J Mol Sci. 2018 Aug 23;19(9):2487. doi: 10.3390/ijms19092487. Int J Mol Sci. 2018. PMID: 30142879 Free PMC article. Review.
-
BMP-7 inhibits TGF-β-induced invasion of breast cancer cells through inhibition of integrin β(3) expression.Cell Oncol (Dordr). 2012 Feb;35(1):19-28. doi: 10.1007/s13402-011-0058-0. Epub 2011 Sep 21. Cell Oncol (Dordr). 2012. PMID: 21935711 Free PMC article.
-
Opposing roles and potential antagonistic mechanism between TGF-β and BMP pathways: Implications for cancer progression.EBioMedicine. 2019 Mar;41:702-710. doi: 10.1016/j.ebiom.2019.02.033. Epub 2019 Feb 23. EBioMedicine. 2019. PMID: 30808576 Free PMC article. Review.
-
Distinct bone morphogenetic proteins activate indistinguishable transcriptional responses in nephron epithelia including Notch _target genes.Cell Signal. 2012 Jan;24(1):257-64. doi: 10.1016/j.cellsig.2011.09.008. Epub 2011 Sep 16. Cell Signal. 2012. PMID: 21945409 Free PMC article.
References
-
- Massague J. How cells read TGF-β signals. Nat Rev Mol Cell Biol. 2000;1:169–178. - PubMed
-
- Morrissey J, Hruska K, Guo G, Wang S, Chen Q, Klahr S. Bone morphogenetic protein-7 improves renal fibrosis and accelerates the return of renal function. J Am Soc Nephrol. 2002;13(Suppl 1):S14–S21. - PubMed
-
- Zeisberg M, Hanai J, Sugimoto H, Mammoto T, Charytan D, Strutz F, Kalluri R. BMP-7 counteracts TGF-β1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med. 2003;9:964–968. - PubMed
-
- Zeisberg M, Bottiglio C, Kumar N, Maeshima Y, Strutz F, Muller GA, Kalluri R. Bone morphogenic protein-7 inhibits progression of chronic renal fibrosis associated with two genetic mouse models. Am J Physiol Renal Physiol. 2003;285:F1060–F1067. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

