Regulation of ERRalpha gene expression by estrogen receptor agonists and antagonists in SKBR3 breast cancer cells: differential molecular mechanisms mediated by g protein-coupled receptor GPR30/GPER-1
- PMID: 20211987
- PMCID: PMC2870941
- DOI: 10.1210/me.2009-0148
Regulation of ERRalpha gene expression by estrogen receptor agonists and antagonists in SKBR3 breast cancer cells: differential molecular mechanisms mediated by g protein-coupled receptor GPR30/GPER-1
Abstract
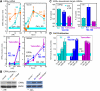
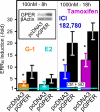
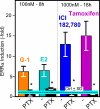
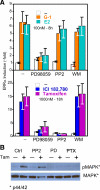
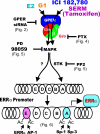
In selected tissues and cell lines, 17beta-estradiol (E2) regulates the expression of estrogen-related receptor alpha (ERRalpha), a member of the orphan nuclear receptor family. This effect is thought to be mediated by the estrogen receptor alpha (ERalpha). However in the ERalpha- and ERbeta-negative SKBR3 breast cancer cell line, physiological levels of E2 also stimulate ERRalpha expression. Here, we explored the molecular mechanism that mediates estrogen action in ER-negative breast cancer cells. We observed that E2, the ERalpha agonist, as well as the ERalpha antagonists ICI 182,780 and tamoxifen (TAM), a selective ER modulator, stimulate the transcriptional activity of the ERRalpha gene and increase the production of ERRalpha protein in SKBR3 cells. Moreover, the ERRalpha downstream _target genes expression and cellular proliferation are also increased. We show further that the G protein-coupled receptor GPR30/GPER-1 (GPER-1) mediates these effects. The GPER-1 specific ligand G-1 mimics the actions of E2, ICI 182,780, and TAM on ERRalpha expression, and changing the levels of GPER-1 mRNA by overexpression or small interfering RNA knockdown affected the expression of ERRalpha accordingly. Utilizing inhibitors, we delineate a different downstream pathway for ER agonist and ER antagonist-triggered signaling through GPER-1. We also find differential histone acetylation and transcription factor recruitment at distinct nucleosomes of the ERRalpha promoter, depending on whether the cells are activated with E2 or with ER antagonists. These findings provide insight into the molecular mechanisms of GPER-1/ERRalpha-mediated signaling and may be relevant to what happens in breast cancer cells escaping inhibitory control by TAM.
Figures


Similar articles
-
G protein-coupled estrogen receptor (GPER) mediates NSCLC progression induced by 17β-estradiol (E2) and selective agonist G1.Med Oncol. 2015 Apr;32(4):104. doi: 10.1007/s12032-015-0558-2. Epub 2015 Mar 6. Med Oncol. 2015. PMID: 25744245
-
Estrogen induces estrogen-related receptor alpha gene expression and chromatin structural changes in estrogen receptor (ER)-positive and ER-negative breast cancer cells.J Biol Chem. 2008 Mar 14;283(11):6752-63. doi: 10.1074/jbc.M705937200. Epub 2008 Jan 2. J Biol Chem. 2008. PMID: 18174157
-
G protein-coupled receptor 30 (GPR30) mediates gene expression changes and growth response to 17beta-estradiol and selective GPR30 ligand G-1 in ovarian cancer cells.Cancer Res. 2007 Feb 15;67(4):1859-66. doi: 10.1158/0008-5472.CAN-06-2909. Cancer Res. 2007. PMID: 17308128
-
[GPER receptor - the new player in estrogen signaling].Postepy Biochem. 2015;61(1):52-60. Postepy Biochem. 2015. PMID: 26281354 Review. Polish.
-
Role of G-protein-coupled estrogen receptor (GPER/GPR30) in maintenance of meiotic arrest in fish oocytes.J Steroid Biochem Mol Biol. 2017 Mar;167:153-161. doi: 10.1016/j.jsbmb.2016.12.005. Epub 2016 Dec 19. J Steroid Biochem Mol Biol. 2017. PMID: 28007532 Review.
Cited by
-
The modulatory role of low concentrations of bisphenol A on tamoxifen-induced proliferation and apoptosis in breast cancer cells.Environ Sci Pollut Res Int. 2019 Jan;26(3):2353-2362. doi: 10.1007/s11356-018-3780-6. Epub 2018 Nov 22. Environ Sci Pollut Res Int. 2019. PMID: 30467747
-
GPR30 as an initiator of tamoxifen resistance in hormone-dependent breast cancer.Breast Cancer Res. 2013 Nov 29;15(6):R114. doi: 10.1186/bcr3581. Breast Cancer Res. 2013. PMID: 24289103 Free PMC article.
-
Estrogen Receptor Is Required for Metformin-Induced Apoptosis in Breast Cancer Cells Under Hyperglycemic Conditions.Breast Cancer (Auckl). 2024 Apr 8;18:11782234241240173. doi: 10.1177/11782234241240173. eCollection 2024. Breast Cancer (Auckl). 2024. PMID: 38779416 Free PMC article.
-
Comparative G-Protein-Coupled Estrogen Receptor (GPER) Systems in Diabetic and Cancer Conditions: A Review.Molecules. 2022 Dec 15;27(24):8943. doi: 10.3390/molecules27248943. Molecules. 2022. PMID: 36558071 Free PMC article. Review.
-
Effects of ICI 182,780, an ERα and ERβ antagonist, and G-1, a GPER agonist, on autophagy in breast cancer cells.Einstein (Sao Paulo). 2020 Apr 22;18:eAO4560. doi: 10.31744/einstein_journal/2020AO4560. eCollection 2020. Einstein (Sao Paulo). 2020. PMID: 32321078 Free PMC article.
References
-
- Yager JD, Davidson NE 2006 Estrogen carcinogenesis in breast cancer. N Engl J Med 354:270–282 - PubMed
-
- Matthews J, Gustafsson JA 2003 Estrogen signaling: a subtle balance between ERα and ERβ. Mol Interv 3:281–292 - PubMed
-
- Klinge CM, Jernigan SC, Mattingly KA, Risinger KE, Zhang J 2004 Estrogen response element-dependent regulation of transcriptional activation of estrogen receptors α and β by coactivators and corepressors. J Mol Endocrinol 33:387–410 - PubMed
-
- Safe S 2001 Transcriptional activation of genes by 17β-estradiol through estrogen receptor-Sp1 interactions. Vitam Horm 62:231–252 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

