Descriptive proteomic analysis shows protein variability between closely related clinical isolates of Mycobacterium tuberculosis
- PMID: 20217870
- PMCID: PMC3517044
- DOI: 10.1002/pmic.200900836
Descriptive proteomic analysis shows protein variability between closely related clinical isolates of Mycobacterium tuberculosis
Abstract
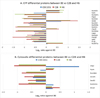
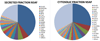
The use of isobaric tags such as iTRAQ allows the relative and absolute quantification of hundreds of proteins in a single experiment for up to eight different samples. More classical techniques such as 2-DE can offer a complimentary approach for the analysis of complex protein samples. In this study, the proteomes of secreted and cytosolic proteins of genetically closely related strains of Mycobacterium tuberculosis were analyzed. Analysis of 2-D gels afforded 28 spots with variations in protein abundance between strains. These were identified by MS/MS. Meanwhile, a rigorous statistical analysis of iTRAQ data allowed the identification and quantification of 101 and 137 proteins in the secreted and cytosolic fractions, respectively. Interestingly, several differences in protein levels were observed between the closely related strains BE, C28 and H6. Seven proteins related to cell wall and cell processes were more abundant in BE, while enzymes related to metabolic pathways (GltA2, SucC, Gnd1, Eno) presented lower levels in the BE strain. Proteins involved in iron and sulfur acquisition (BfrB, ViuB, TB15.3 and SseC2) were more abundant in C28 and H6. In general, iTRAQ afforded rapid identification of fine differences between protein levels such as those presented between closely related strains. This provides a platform from which the relevance of these differences can be assessed further using complimentary proteomic and biological modeling methods.
Conflict of interest statement
All of the authors declare no conflicts of interest.
Figures

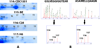
Similar articles
-
Mycobacterium tuberculosis in the Proteomics Era.Microbiol Spectr. 2014 Apr;2(2). doi: 10.1128/microbiolspec.MGM2-0020-2013. Microbiol Spectr. 2014. PMID: 26105825 Review.
-
[Quantitative proteomic analysis of streptomycin resistant and sensitive clinical isolates of Mycobacterium tuberculosis].Wei Sheng Wu Xue Bao. 2013 Feb 4;53(2):154-63. Wei Sheng Wu Xue Bao. 2013. PMID: 23627108 Chinese.
-
Proteomics technologies for the global identification and quantification of proteins.Adv Protein Chem Struct Biol. 2010;80:1-44. doi: 10.1016/B978-0-12-381264-3.00001-1. Adv Protein Chem Struct Biol. 2010. PMID: 21109216 Review.
-
Comparative proteomics analysis between biofilm and planktonic cells of Mycobacterium tuberculosis.Electrophoresis. 2019 Oct;40(20):2736-2746. doi: 10.1002/elps.201900030. Epub 2019 Jun 6. Electrophoresis. 2019. PMID: 31141184
-
What Can Proteomics Tell Us about Tuberculosis?J Microbiol Biotechnol. 2015 Aug;25(8):1181-94. doi: 10.4014/jmb.1502.02008. J Microbiol Biotechnol. 2015. PMID: 25737117 Review.
Cited by
-
Identification and architecture of a putative secretion tube across mycobacterial outer envelope.Sci Adv. 2021 Aug 20;7(34):eabg5656. doi: 10.1126/sciadv.abg5656. Print 2021 Aug. Sci Adv. 2021. PMID: 34417177 Free PMC article.
-
Upregulation of the phthiocerol dimycocerosate biosynthetic pathway by rifampin-resistant, rpoB mutant Mycobacterium tuberculosis.J Bacteriol. 2012 Dec;194(23):6441-52. doi: 10.1128/JB.01013-12. Epub 2012 Sep 21. J Bacteriol. 2012. PMID: 23002228 Free PMC article.
-
Deciphering the proteome of the in vivo diagnostic reagent "purified protein derivative" from Mycobacterium tuberculosis.Proteomics. 2012 Apr;12(7):979-91. doi: 10.1002/pmic.201100544. Proteomics. 2012. PMID: 22522804 Free PMC article.
-
Proteomics for the Investigation of Mycobacteria.Acta Naturae. 2017 Jan-Mar;9(1):15-25. Acta Naturae. 2017. PMID: 28461970 Free PMC article.
-
Epidemiologic consequences of microvariation in Mycobacterium tuberculosis.J Infect Dis. 2012 Mar 15;205(6):964-74. doi: 10.1093/infdis/jir876. Epub 2012 Feb 7. J Infect Dis. 2012. PMID: 22315279 Free PMC article.
References
-
- Ross PL, Huang YN, Marchese JN, Williamson B, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. - PubMed
-
- D'Ascenzo M, Choe L, Lee KH. iTRAQPak: an R based analysis and visualization package for 8-plex isobaric protein expression data. Brief Funct Genomic Proteomic. 2008;7:127–135. - PubMed
-
- Chen Y, Choong LY, Lin Q, Philp R, et al. Differential expression of novel tyrosine kinase substrates during breast cancer development. Mol Cell Proteomics. 2007;6:2072–2087. - PubMed
-
- Drummelsmith J, Winstall E, Bergeron MG, Poirier GG, Ouellette M. Comparative proteomics analyses reveal a potential biomarker for the detection of vancomycin-intermediate Staphylococcus aureus strains. J Proteome Res. 2007;6:4690–4702. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

