A rapid and quantitative LC-MS/MS method to profile sphingolipids
- PMID: 20228220
- PMCID: PMC2882728
- DOI: 10.1194/jlr.D005322
A rapid and quantitative LC-MS/MS method to profile sphingolipids
Abstract
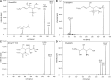
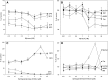
Sphingolipids comprise a highly diverse and complex class of molecules that serve not only as structural components of membranes but also as signaling molecules. To understand the differential role of sphingolipids in a regulatory network, it is important to use specific and quantitative methods. We developed a novel LC-MS/MS method for the rapid, simultaneous quantification of sphingolipid metabolites, including sphingosine, sphinganine, phyto-sphingosine, di- and trimethyl-sphingosine, sphingosylphosphorylcholine, hexosylceramide, lactosylceramide, ceramide-1-phosphate, and dihydroceramide-1-phosphate. Appropriate internal standards (ISs) were added prior to lipid extraction. In contrast to most published methods based on reversed phase chromatography, we used hydrophilic interaction liquid chromatography and achieved good peak shapes, a short analysis time of 4.5 min, and, most importantly, coelution of analytes and their respective ISs. To avoid an overestimation of species concentrations, peak areas were corrected regarding isotopic overlap where necessary. Quantification was achieved by standard addition of naturally occurring sphingolipid species to the sample matrix. The method showed excellent precision, accuracy, detection limits, and robustness. As an example, sphingolipid species were quantified in fibroblasts treated with myriocin or sphingosine-kinase inhibitor. In summary, this method represents a valuable tool to evaluate the role of sphingolipids in the regulation of cell functions.
Figures


Similar articles
-
Structure-specific, quantitative methods for analysis of sphingolipids by liquid chromatography-tandem mass spectrometry: "inside-out" sphingolipidomics.Methods Enzymol. 2007;432:83-115. doi: 10.1016/S0076-6879(07)32004-1. Methods Enzymol. 2007. PMID: 17954214
-
A simple and rapid method for extraction and measurement of circulating sphingolipids using LC-MS/MS: a _targeted lipidomic analysis.Anal Bioanal Chem. 2022 Mar;414(6):2041-2054. doi: 10.1007/s00216-021-03853-z. Epub 2022 Jan 23. Anal Bioanal Chem. 2022. PMID: 35066602
-
Sphingolipid profiling of human plasma and FPLC-separated lipoprotein fractions by hydrophilic interaction chromatography tandem mass spectrometry.Biochim Biophys Acta. 2011 Feb;1811(2):68-75. doi: 10.1016/j.bbalip.2010.11.003. Epub 2010 Nov 23. Biochim Biophys Acta. 2011. PMID: 21081176
-
Sphingolipid analysis by high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS).Adv Exp Med Biol. 2010;688:46-59. doi: 10.1007/978-1-4419-6741-1_3. Adv Exp Med Biol. 2010. PMID: 20919645 Review.
-
Analysis of mammalian sphingolipids by liquid chromatography tandem mass spectrometry (LC-MS/MS) and tissue imaging mass spectrometry (TIMS).Biochim Biophys Acta. 2011 Nov;1811(11):838-53. doi: 10.1016/j.bbalip.2011.06.027. Epub 2011 Jul 1. Biochim Biophys Acta. 2011. PMID: 21749933 Free PMC article. Review.
Cited by
-
Comparative lipidomic profiling of the human commensal bacterium Propionibacterium acnes and its extracellular vesicles.RSC Adv. 2018 Apr 23;8(27):15241-15247. doi: 10.1039/c7ra13769a. eCollection 2018 Apr 18. RSC Adv. 2018. PMID: 35541326 Free PMC article.
-
Enhancement of human plasma glucosylceramide assay sensitivity using delipidized plasma.Mol Genet Metab Rep. 2016 Aug 7;8:77-9. doi: 10.1016/j.ymgmr.2016.07.004. eCollection 2016 Sep. Mol Genet Metab Rep. 2016. PMID: 27547732 Free PMC article.
-
Deciphering sphingolipid biosynthesis dynamics in Arabidopsis thaliana cell cultures: Quantitative analysis amid data variability.iScience. 2024 Aug 5;27(9):110675. doi: 10.1016/j.isci.2024.110675. eCollection 2024 Sep 20. iScience. 2024. PMID: 39297170 Free PMC article.
-
Functional Insights into the Sphingolipids C1P, S1P, and SPC in Human Fibroblast-like Synoviocytes by Proteomic Analysis.Int J Mol Sci. 2024 Jul 31;25(15):8363. doi: 10.3390/ijms25158363. Int J Mol Sci. 2024. PMID: 39125932 Free PMC article.
-
Comparison of Workflows for Milk Lipid Analysis: Phospholipids.Foods. 2022 Dec 28;12(1):163. doi: 10.3390/foods12010163. Foods. 2022. PMID: 36613379 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

