Artificial and natural duplicates in pyrosequencing reads of metagenomic data
- PMID: 20388221
- PMCID: PMC2874554
- DOI: 10.1186/1471-2105-11-187
Artificial and natural duplicates in pyrosequencing reads of metagenomic data
Abstract
Background: Artificial duplicates from pyrosequencing reads may lead to incorrect interpretation of the abundance of species and genes in metagenomic studies. Duplicated reads were filtered out in many metagenomic projects. However, since the duplicated reads observed in a pyrosequencing run also include natural (non-artificial) duplicates, simply removing all duplicates may also cause underestimation of abundance associated with natural duplicates.
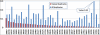
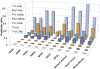
Results: We implemented a method for identification of exact and nearly identical duplicates from pyrosequencing reads. This method performs an all-against-all sequence comparison and clusters the duplicates into groups using an algorithm modified from our previous sequence clustering method cd-hit. This method can process a typical dataset in approximately 10 minutes; it also provides a consensus sequence for each group of duplicates. We applied this method to the underlying raw reads of 39 genomic projects and 10 metagenomic projects that utilized pyrosequencing technique. We compared the occurrences of the duplicates identified by our method and the natural duplicates made by independent simulations. We observed that the duplicates, including both artificial and natural duplicates, make up 4-44% of reads. The number of natural duplicates highly correlates with the samples' read density (number of reads divided by genome size). For high-complexity metagenomic samples lacking dominant species, natural duplicates only make up <1% of all duplicates. But for some other samples like transcriptomic samples, majority of the observed duplicates might be natural duplicates.
Conclusions: Our method is available from http://cd-hit.org as a downloadable program and a web server. It is important not only to identify the duplicates from metagenomic datasets but also to distinguish whether they are artificial or natural duplicates. We provide a tool to estimate the number of natural duplicates according to user-defined sample types, so users can decide whether to retain or remove duplicates in their projects.
Figures
Similar articles
-
Selection of marker genes for genetic barcoding of microorganisms and binning of metagenomic reads by Barcoder software tools.BMC Bioinformatics. 2018 Aug 30;19(1):309. doi: 10.1186/s12859-018-2320-1. BMC Bioinformatics. 2018. PMID: 30165813 Free PMC article.
-
TagCleaner: Identification and removal of tag sequences from genomic and metagenomic datasets.BMC Bioinformatics. 2010 Jun 23;11:341. doi: 10.1186/1471-2105-11-341. BMC Bioinformatics. 2010. PMID: 20573248 Free PMC article.
-
Identifying and removing artificial replicates from 454 pyrosequencing data.Cold Spring Harb Protoc. 2010 Apr;2010(4):pdb.prot5409. doi: 10.1101/pdb.prot5409. Cold Spring Harb Protoc. 2010. PMID: 20360363
-
FR-HIT, a very fast program to recruit metagenomic reads to homologous reference genomes.Bioinformatics. 2011 Jun 15;27(12):1704-5. doi: 10.1093/bioinformatics/btr252. Epub 2011 Apr 19. Bioinformatics. 2011. PMID: 21505035 Free PMC article.
-
Assessment of metagenomic assemblers based on hybrid reads of real and simulated metagenomic sequences.Brief Bioinform. 2020 May 21;21(3):777-790. doi: 10.1093/bib/bbz025. Brief Bioinform. 2020. PMID: 30860572 Free PMC article. Review.
Cited by
-
Unbiased approach for virus detection in skin lesions.PLoS One. 2013 Jun 28;8(6):e65953. doi: 10.1371/journal.pone.0065953. Print 2013. PLoS One. 2013. PMID: 23840382 Free PMC article.
-
Effect of polybrominated diphenyl ether (PBDE) treatment on the composition and function of the bacterial community in the sponge Haliclona cymaeformis.Front Microbiol. 2015 Jan 14;5:799. doi: 10.3389/fmicb.2014.00799. eCollection 2014. Front Microbiol. 2015. PMID: 25642227 Free PMC article.
-
A multilabel model based on Chou's pseudo-amino acid composition for identifying membrane proteins with both single and multiple functional types.J Membr Biol. 2013 Apr;246(4):327-34. doi: 10.1007/s00232-013-9536-9. Epub 2013 Apr 2. J Membr Biol. 2013. PMID: 23546013
-
The Pacific Ocean virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology.PLoS One. 2013;8(2):e57355. doi: 10.1371/journal.pone.0057355. Epub 2013 Feb 28. PLoS One. 2013. PMID: 23468974 Free PMC article.
-
PyroTRF-ID: a novel bioinformatics methodology for the affiliation of terminal-restriction fragments using 16S rRNA gene pyrosequencing data.BMC Microbiol. 2012 Dec 27;12:306. doi: 10.1186/1471-2180-12-306. BMC Microbiol. 2012. PMID: 23270314 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

