Inhibition of hepatitis B virus replication by MyD88 involves accelerated degradation of pregenomic RNA and nuclear retention of pre-S/S RNAs
- PMID: 20410269
- PMCID: PMC2903248
- DOI: 10.1128/JVI.00236-10
Inhibition of hepatitis B virus replication by MyD88 involves accelerated degradation of pregenomic RNA and nuclear retention of pre-S/S RNAs
Abstract
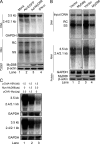
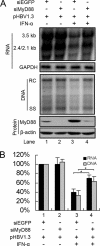
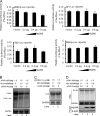
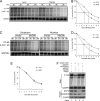
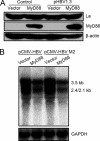
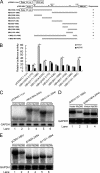
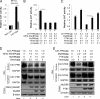
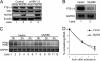
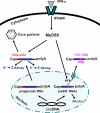
Myeloid differentiation primary response protein 88 (MyD88), which can be induced by alpha interferon (IFN-alpha), has an antiviral activity against the hepatitis B virus (HBV). The mechanism of this antiviral activity remains poorly understood. Here, we report that MyD88 inhibited HBV replication in HepG2.2.15 cells and in a mouse model. The knockdown of MyD88 expression weakened the IFN-alpha-induced inhibition of HBV replication. Furthermore, MyD88 posttranscriptionally reduced the levels of viral RNA. Remarkably, MyD88 accelerated the decay of viral pregenomic RNA in the cytoplasm. Mapping analysis showed that the RNA sequence located in the 5'-proximal region of the pregenomic RNA was critical for the decay. In addition, MyD88 inhibited the nuclear export of pre-S/S RNAs via the posttranscriptional regulatory element (PRE). The retained pre-S/S RNAs were shown to degrade in the nucleus. Finally, we found that MyD88 inhibited the expression of polypyrimidine tract-binding protein (PTB), a key nuclear export factor for PRE-containing RNA. Taken together, our results define a novel antiviral mechanism against HBV mediated by MyD88.
Figures

Similar articles
-
Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein.PLoS Pathog. 2013;9(7):e1003494. doi: 10.1371/journal.ppat.1003494. Epub 2013 Jul 11. PLoS Pathog. 2013. PMID: 23853601 Free PMC article.
-
NgAgo-gDNA system efficiently suppresses hepatitis B virus replication through accelerating decay of pregenomic RNA.Antiviral Res. 2017 Sep;145:20-23. doi: 10.1016/j.antiviral.2017.07.005. Epub 2017 Jul 12. Antiviral Res. 2017. PMID: 28709658
-
Inhibition of hepatitis B virus replication by cIAP2 involves accelerating the ubiquitin-proteasome-mediated destruction of polymerase.J Virol. 2011 Nov;85(21):11457-67. doi: 10.1128/JVI.00879-11. Epub 2011 Aug 24. J Virol. 2011. PMID: 21865390 Free PMC article.
-
Activation of pattern recognition receptor-mediated innate immunity inhibits the replication of hepatitis B virus in human hepatocyte-derived cells.J Virol. 2009 Jan;83(2):847-58. doi: 10.1128/JVI.02008-08. Epub 2008 Oct 29. J Virol. 2009. PMID: 18971270 Free PMC article.
-
Interferon-inducible MyD88 protein inhibits hepatitis B virus replication.Virology. 2004 Feb 20;319(2):306-14. doi: 10.1016/j.virol.2003.11.005. Virology. 2004. PMID: 14980490
Cited by
-
The Interferon-Inducible Protein Tetherin Inhibits Hepatitis B Virus Virion Secretion.J Virol. 2015 Sep;89(18):9200-12. doi: 10.1128/JVI.00933-15. Epub 2015 Jun 24. J Virol. 2015. PMID: 26109732 Free PMC article.
-
Inhibition of hepatitis B virus gene expression and replication by hepatocyte nuclear factor 6.J Virol. 2015 Apr;89(8):4345-55. doi: 10.1128/JVI.03094-14. Epub 2015 Feb 4. J Virol. 2015. PMID: 25653429 Free PMC article.
-
Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment.J Virol. 2011 Jul;85(13):6319-33. doi: 10.1128/JVI.02627-10. Epub 2011 Apr 20. J Virol. 2011. PMID: 21507968 Free PMC article.
-
Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity.Nat Immunol. 2013 Aug;14(8):793-803. doi: 10.1038/ni.2647. Epub 2013 Jul 7. Nat Immunol. 2013. PMID: 23832071
-
Innate Immunity, Inflammation, and Intervention in HBV Infection.Viruses. 2022 Oct 17;14(10):2275. doi: 10.3390/v14102275. Viruses. 2022. PMID: 36298831 Free PMC article. Review.
References
-
- Bonvin, M., F. Achermann, I. Greeve, D. Stroka, A. Keogh, D. Inderbitzin, D. Candinas, P. Sommer, S. Wain-Hobson, J. P. Vartanian, and J. Greeve. 2006. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 43:1364-1374. - PubMed
-
- Chebath, J., P. Benech, M. Revel, and M. Vigneron. 1987. Constitutive expression of (2′-5′) oligo A synthetase confers resistance to picornavirus infection. Nature 330:587-588. - PubMed
-
- Dienstag, J. L. 2008. Hepatitis B virus infection. N. Engl. J. Med. 359:1486-1500. - PubMed
-
- Ehlers, I., S. Horke, K. Reumann, A. Rang, F. Grosse, H. Will, and T. Heise. 2004. Functional characterization of the interaction between human La and hepatitis B virus RNA. J. Biol. Chem. 279:43437-43447. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

