TumorBoost: normalization of allele-specific tumor copy numbers from a single pair of tumor-normal genotyping microarrays
- PMID: 20462408
- PMCID: PMC2894037
- DOI: 10.1186/1471-2105-11-245
TumorBoost: normalization of allele-specific tumor copy numbers from a single pair of tumor-normal genotyping microarrays
Abstract
Background: High-throughput genotyping microarrays assess both total DNA copy number and allelic composition, which makes them a tool of choice for copy number studies in cancer, including total copy number and loss of heterozygosity (LOH) analyses. Even after state of the art preprocessing methods, allelic signal estimates from genotyping arrays still suffer from systematic effects that make them difficult to use effectively for such downstream analyses.
Results: We propose a method, TumorBoost, for normalizing allelic estimates of one tumor sample based on estimates from a single matched normal. The method applies to any paired tumor-normal estimates from any microarray-based technology, combined with any preprocessing method. We demonstrate that it increases the signal-to-noise ratio of allelic signals, making it significantly easier to detect allelic imbalances.
Conclusions: TumorBoost increases the power to detect somatic copy-number events (including copy-neutral LOH) in the tumor from allelic signals of Affymetrix or Illumina origin. We also conclude that high-precision allelic estimates can be obtained from a single pair of tumor-normal hybridizations, if TumorBoost is combined with single-array preprocessing methods such as (allele-specific) CRMA v2 for Affymetrix or BeadStudio's (proprietary) XY-normalization method for Illumina. A bounded-memory implementation is available in the open-source and cross-platform R package aroma.cn, which is part of the Aroma Project (http://www.aroma-project.org/).
Figures


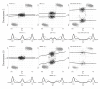
 ), and empirical densities of the raw (βT; dashed) and the normalized (
), and empirical densities of the raw (βT; dashed) and the normalized ( ; solid) allele B fractions for sample TCGA-23-1027. The same regions, SNPs and annotation as in Figure 2 are used.
; solid) allele B fractions for sample TCGA-23-1027. The same regions, SNPs and annotation as in Figure 2 are used.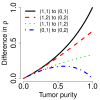

 ) heterozygous DHs, respectively. A 1000 kb safety region (dashed gray frame) around the change point is excluded from the evaluation. The full resolution data points are colored black and the binned (H = 4) ones are colored blue. The three panels in the bottom row show the ROC performance of the TCNs (dotted green) and the raw (dashed black) and normalized (solid red and dot-dashed blue for naive and population-based genotypes, respectively) DHs at the full resolution (H = 1; no binning), and after binning in non-overlapping windows of size H = 2 and H = 4 SNPs, respectively.
) heterozygous DHs, respectively. A 1000 kb safety region (dashed gray frame) around the change point is excluded from the evaluation. The full resolution data points are colored black and the binned (H = 4) ones are colored blue. The three panels in the bottom row show the ROC performance of the TCNs (dotted green) and the raw (dashed black) and normalized (solid red and dot-dashed blue for naive and population-based genotypes, respectively) DHs at the full resolution (H = 1; no binning), and after binning in non-overlapping windows of size H = 2 and H = 4 SNPs, respectively.

Similar articles
-
CalMaTe: a method and software to improve allele-specific copy number of SNP arrays for downstream segmentation.Bioinformatics. 2012 Jul 1;28(13):1793-4. doi: 10.1093/bioinformatics/bts248. Epub 2012 May 9. Bioinformatics. 2012. PMID: 22576175 Free PMC article.
-
Normalization of Illumina Infinium whole-genome SNP data improves copy number estimates and allelic intensity ratios.BMC Bioinformatics. 2008 Oct 2;9:409. doi: 10.1186/1471-2105-9-409. BMC Bioinformatics. 2008. PMID: 18831757 Free PMC article.
-
Estimation and assessment of raw copy numbers at the single locus level.Bioinformatics. 2008 Mar 15;24(6):759-67. doi: 10.1093/bioinformatics/btn016. Epub 2008 Jan 19. Bioinformatics. 2008. PMID: 18204055
-
Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotide-polymorphism genotyping microarrays.Am J Hum Genet. 2007 Jul;81(1):114-26. doi: 10.1086/518809. Epub 2007 Jun 5. Am J Hum Genet. 2007. PMID: 17564968 Free PMC article.
-
Single-nucleotide polymorphism array karyotyping in clinical practice: where, when, and how?Semin Oncol. 2012 Feb;39(1):13-25. doi: 10.1053/j.seminoncol.2011.11.010. Semin Oncol. 2012. PMID: 22289488 Review.
Cited by
-
CalMaTe: a method and software to improve allele-specific copy number of SNP arrays for downstream segmentation.Bioinformatics. 2012 Jul 1;28(13):1793-4. doi: 10.1093/bioinformatics/bts248. Epub 2012 May 9. Bioinformatics. 2012. PMID: 22576175 Free PMC article.
-
Abundant copy-number loss of CYCLOPS and STOP genes in gastric adenocarcinoma.Gastric Cancer. 2016 Apr;19(2):453-465. doi: 10.1007/s10120-015-0514-z. Epub 2015 Jul 24. Gastric Cancer. 2016. PMID: 26205786 Free PMC article.
-
MethylPurify: tumor purity deconvolution and differential methylation detection from single tumor DNA methylomes.Genome Biol. 2014 Aug 7;15(8):419. doi: 10.1186/s13059-014-0419-x. Genome Biol. 2014. PMID: 25103624 Free PMC article.
-
Estimation of parent specific DNA copy number in tumors using high-density genotyping arrays.PLoS Comput Biol. 2011 Jan 27;7(1):e1001060. doi: 10.1371/journal.pcbi.1001060. PLoS Comput Biol. 2011. PMID: 21298078 Free PMC article.
-
AbsCN-seq: a statistical method to estimate tumor purity, ploidy and absolute copy numbers from next-generation sequencing data.Bioinformatics. 2014 Apr 15;30(8):1056-1063. doi: 10.1093/bioinformatics/btt759. Epub 2014 Jan 2. Bioinformatics. 2014. PMID: 24389661 Free PMC article.
References
-
- Affymetrix Inc. Genome-Wide Human SNP Nsp/Sty 6.0 user guide. Affymetrix Inc; 2007. [Rev 1.]
-
- Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F, Haden K, Li J, Shaw CA, Belmont J, Cheung SW, Shen RM, Barker DL, Gunderson KL. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16(9):1136–1148. doi: 10.1101/gr.5402306. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

