Alu sequences in undifferentiated human embryonic stem cells display high levels of A-to-I RNA editing
- PMID: 20574523
- PMCID: PMC2888580
- DOI: 10.1371/journal.pone.0011173
Alu sequences in undifferentiated human embryonic stem cells display high levels of A-to-I RNA editing
Abstract
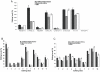
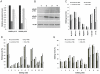
Adenosine to Inosine (A-to-I) RNA editing is a site-specific modification of RNA transcripts, catalyzed by members of the ADAR (Adenosine Deaminase Acting on RNA) protein family. RNA editing occurs in human RNA in thousands of different sites. Some of the sites are located in protein-coding regions but the majority is found in non-coding regions, such as 3'UTRs, 5'UTRs and introns - mainly in Alu elements. While editing is found in all tissues, the highest levels of editing are found in the brain. It was shown that editing levels within protein-coding regions are increased during embryogenesis and after birth and that RNA editing is crucial for organism viability as well as for normal development. In this study we characterized the A-to-I RNA editing phenomenon during neuronal and spontaneous differentiation of human embryonic stem cells (hESCs). We identified high editing levels of Alu repetitive elements in hESCs and demonstrated a global decrease in editing levels of non-coding Alu sites when hESCs are differentiating, particularly into the neural lineage. Using RNA interference, we showed that the elevated editing levels of Alu elements in undifferentiated hESCs are highly dependent on ADAR1. DNA microarray analysis showed that ADAR1 knockdown has a global effect on gene expression in hESCs and leads to a significant increase in RNA expression levels of genes involved in differentiation and development processes, including neurogenesis. Taken together, we speculate that A-to-I editing of Alu sequences plays a role in the regulation of hESC early differentiation decisions.
Conflict of interest statement
Figures
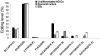
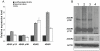
Similar articles
-
Altered A-to-I RNA editing in human embryogenesis.PLoS One. 2012;7(7):e41576. doi: 10.1371/journal.pone.0041576. Epub 2012 Jul 31. PLoS One. 2012. PMID: 22859999 Free PMC article.
-
Adenosine-to-inosine Alu RNA editing controls the stability of the pro-inflammatory long noncoding RNA NEAT1 in atherosclerotic cardiovascular disease.J Mol Cell Cardiol. 2021 Nov;160:111-120. doi: 10.1016/j.yjmcc.2021.07.005. Epub 2021 Jul 21. J Mol Cell Cardiol. 2021. PMID: 34302813 Free PMC article.
-
A-to-I editing of coding and non-coding RNAs by ADARs.Nat Rev Mol Cell Biol. 2016 Feb;17(2):83-96. doi: 10.1038/nrm.2015.4. Epub 2015 Dec 9. Nat Rev Mol Cell Biol. 2016. PMID: 26648264 Free PMC article. Review.
-
ADAR1 is involved in the regulation of reprogramming human fibroblasts to induced pluripotent stem cells.Stem Cells Dev. 2014 Mar 1;23(5):443-56. doi: 10.1089/scd.2013.0206. Epub 2013 Dec 14. Stem Cells Dev. 2014. PMID: 24192045
-
All I's on the RADAR: role of ADAR in gene regulation.FEBS Lett. 2018 Sep;592(17):2860-2873. doi: 10.1002/1873-3468.13093. Epub 2018 May 25. FEBS Lett. 2018. PMID: 29770436 Free PMC article. Review.
Cited by
-
Circular RNAs in renal cell carcinoma: from mechanistic to clinical perspective.Cancer Cell Int. 2023 Nov 22;23(1):288. doi: 10.1186/s12935-023-03128-w. Cancer Cell Int. 2023. PMID: 37993909 Free PMC article. Review.
-
Epitranscriptomic modifications in mesenchymal stem cell differentiation: advances, mechanistic insights, and beyond.Cell Death Differ. 2024 Jan;31(1):9-27. doi: 10.1038/s41418-023-01238-6. Epub 2023 Nov 20. Cell Death Differ. 2024. PMID: 37985811 Review.
-
Emerging role of the RNA-editing enzyme ADAR1 in stem cell fate and function.Biomark Res. 2023 Jun 6;11(1):61. doi: 10.1186/s40364-023-00503-7. Biomark Res. 2023. PMID: 37280687 Free PMC article. Review.
-
FUS Alters circRNA Metabolism in Human Motor Neurons Carrying the ALS-Linked P525L Mutation.Int J Mol Sci. 2023 Feb 6;24(4):3181. doi: 10.3390/ijms24043181. Int J Mol Sci. 2023. PMID: 36834591 Free PMC article.
-
Specific expression and functions of circular RNAs.Cell Death Differ. 2022 Mar;29(3):481-491. doi: 10.1038/s41418-022-00948-7. Epub 2022 Feb 15. Cell Death Differ. 2022. PMID: 35169296 Free PMC article. Review.
References
-
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. - PubMed
-
- Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. - PubMed
-
- Dvash T, Mayshar Y, Darr H, McElhaney M, Barker D, et al. Temporal gene expression during differentiation of human embryonic stem cells and embryoid bodies. Hum Reprod. 2004;19:2875–2883. - PubMed
-
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

