Prion amyloid structure explains templating: how proteins can be genes
- PMID: 20726897
- PMCID: PMC3025496
- DOI: 10.1111/j.1567-1364.2010.00666.x
Prion amyloid structure explains templating: how proteins can be genes
Abstract
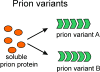
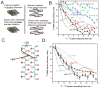
The yeast and fungal prions determine heritable and infectious traits, and are thus genes composed of protein. Most prions are inactive forms of a normal protein as it forms a self-propagating filamentous β-sheet-rich polymer structure called amyloid. Remarkably, a single prion protein sequence can form two or more faithfully inherited prion variants, in effect alleles of these genes. What protein structure explains this protein-based inheritance? Using solid-state nuclear magnetic resonance, we showed that the infectious amyloids of the prion domains of Ure2p, Sup35p and Rnq1p have an in-register parallel architecture. This structure explains how the amyloid filament ends can template the structure of a new protein as it joins the filament. The yeast prions [PSI(+)] and [URE3] are not found in wild strains, indicating that they are a disadvantage to the cell. Moreover, the prion domains of Ure2p and Sup35p have functions unrelated to prion formation, indicating that these domains are not present for the purpose of forming prions. Indeed, prion-forming ability is not conserved, even within Saccharomyces cerevisiae, suggesting that the rare formation of prions is a disease. The prion domain sequences generally vary more rapidly in evolution than does the remainder of the molecule, producing a barrier to prion transmission, perhaps selected in evolution by this protection.
Journal compilation © 2010 Federation of European Microbiological Societies. Published by Blackwell Publishing Ltd. No claim to original US government works.
Figures


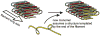
Similar articles
-
Primary sequence independence for prion formation.Proc Natl Acad Sci U S A. 2005 Sep 6;102(36):12825-30. doi: 10.1073/pnas.0506136102. Epub 2005 Aug 25. Proc Natl Acad Sci U S A. 2005. PMID: 16123127 Free PMC article.
-
Prions are affected by evolution at two levels.Cell Mol Life Sci. 2016 Mar;73(6):1131-44. doi: 10.1007/s00018-015-2109-6. Epub 2015 Dec 28. Cell Mol Life Sci. 2016. PMID: 26713322 Free PMC article. Review.
-
Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register beta-sheet structure.Proc Natl Acad Sci U S A. 2008 Feb 19;105(7):2403-8. doi: 10.1073/pnas.0712032105. Epub 2008 Feb 11. Proc Natl Acad Sci U S A. 2008. PMID: 18268327 Free PMC article.
-
Protein inheritance (prions) based on parallel in-register beta-sheet amyloid structures.Bioessays. 2008 Oct;30(10):955-64. doi: 10.1002/bies.20821. Bioessays. 2008. PMID: 18798523 Free PMC article. Review.
-
A promiscuous prion: efficient induction of [URE3] prion formation by heterologous prion domains.Genetics. 2009 Nov;183(3):929-40. doi: 10.1534/genetics.109.109322. Epub 2009 Sep 14. Genetics. 2009. PMID: 19752212 Free PMC article.
Cited by
-
Discovering protein-based inheritance through yeast genetics.J Biol Chem. 2012 Apr 27;287(18):14432-42. doi: 10.1074/jbc.X112.355636. Epub 2012 Mar 6. J Biol Chem. 2012. PMID: 22396539 Free PMC article. Review. No abstract available.
-
Mutations Outside the Ure2 Amyloid-Forming Region Disrupt [URE3] Prion Propagation and Alter Interactions with Protein Quality Control Factors.Mol Cell Biol. 2020 Oct 13;40(21):e00294-20. doi: 10.1128/MCB.00294-20. Print 2020 Oct 13. Mol Cell Biol. 2020. PMID: 32868289 Free PMC article.
-
[PSI+] maintenance is dependent on the composition, not primary sequence, of the oligopeptide repeat domain.PLoS One. 2011;6(7):e21953. doi: 10.1371/journal.pone.0021953. Epub 2011 Jul 8. PLoS One. 2011. PMID: 21760933 Free PMC article.
-
Amyloid and the origin of life: self-replicating catalytic amyloids as prebiotic informational and protometabolic entities.Cell Mol Life Sci. 2018 May;75(9):1499-1507. doi: 10.1007/s00018-018-2797-9. Epub 2018 Mar 17. Cell Mol Life Sci. 2018. PMID: 29550973 Free PMC article.
-
Prion protein at the crossroads of physiology and disease.Trends Neurosci. 2012 Feb;35(2):92-103. doi: 10.1016/j.tins.2011.10.002. Epub 2011 Dec 1. Trends Neurosci. 2012. PMID: 22137337 Free PMC article. Review.
References
-
- Alper T, Haig DA, Clarke MC. The exceptionally small size of the scrapie agent. Biochem Biophys Res Commun. 1966;22:278 – 284. - PubMed
-
- Alper T, Cramp WA, Haig DA, Clarke MC. Does the agent of scrapie replicate without nucleic acid? Nature. 1967;214:764 – 766. - PubMed
-
- Bai M, Zhou JM, Perrett S. The yeast prion protein Ure2 shows glutathione peroxidase activity in both native and fibrillar forms. J Biol Chem. 2004;279:50025–50030. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

