MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing
- PMID: 20808952
- PMCID: PMC2923083
- DOI: 10.1371/journal.pbio.1000453
MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing
Abstract
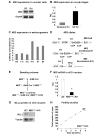
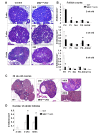
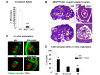
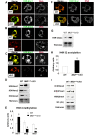
During gametogenesis and pre-implantation development, the mammalian epigenome is reprogrammed to establish pluripotency in the epiblast. Here we show that the histone 3 lysine 4 (H3K4) methyltransferase, MLL2, controls most of the promoter-specific chromatin modification, H3K4me3, during oogenesis and early development. Using conditional knockout mutagenesis and a hypomorph model, we show that Mll2 deficiency in oocytes results in anovulation and oocyte death, with increased transcription of p53, apoptotic factors, and Iap elements. MLL2 is required for (1) bulk H3K4me3 but not H3K4me1, indicating that MLL2 controls most promoters but monomethylation is regulated by a different H3K4 methyltransferase; (2) the global transcriptional silencing that preceeds resumption of meiosis but not for the concomitant nuclear reorganization into the surrounded nucleolus (SN) chromatin configuration; (3) oocyte survival; and (4) normal zygotic genome activation. These results reveal that MLL2 is autonomously required in oocytes for fertility and imply that MLL2 contributes to the epigenetic reprogramming that takes place before fertilization. We propose that once this task has been accomplished, MLL2 is not required until gastrulation and that other methyltransferases are responsible for bulk H3K4me3, thereby revealing an unexpected epigenetic control switch amongst the H3K4 methyltransferases during development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant.Development. 2014 Feb;141(3):526-37. doi: 10.1242/dev.102681. Epub 2014 Jan 14. Development. 2014. PMID: 24423662
-
MLL2 conveys transcription-independent H3K4 trimethylation in oocytes.Nat Struct Mol Biol. 2018 Jan;25(1):73-82. doi: 10.1038/s41594-017-0013-5. Epub 2018 Jan 1. Nat Struct Mol Biol. 2018. PMID: 29323282
-
Uncoupling histone H3K4 trimethylation from developmental gene expression via an equilibrium of COMPASS, Polycomb and DNA methylation.Nat Genet. 2020 Jun;52(6):615-625. doi: 10.1038/s41588-020-0618-1. Epub 2020 May 11. Nat Genet. 2020. PMID: 32393859 Free PMC article.
-
SET/MLL family proteins in hematopoiesis and leukemia.Int J Hematol. 2017 Jan;105(1):7-16. doi: 10.1007/s12185-016-2118-8. Epub 2016 Oct 31. Int J Hematol. 2017. PMID: 27796741 Review.
-
Structure, Activity and Function of the MLL2 (KMT2B) Protein Lysine Methyltransferase.Life (Basel). 2021 Aug 12;11(8):823. doi: 10.3390/life11080823. Life (Basel). 2021. PMID: 34440566 Free PMC article. Review.
Cited by
-
KMT2 Family of H3K4 Methyltransferases: Enzymatic Activity-dependent and -independent Functions.J Mol Biol. 2024 Apr 1;436(7):168453. doi: 10.1016/j.jmb.2024.168453. Epub 2024 Jan 22. J Mol Biol. 2024. PMID: 38266981 Review.
-
Insights into epigenetic patterns in mammalian early embryos.Protein Cell. 2021 Jan;12(1):7-28. doi: 10.1007/s13238-020-00757-z. Epub 2020 Jul 15. Protein Cell. 2021. PMID: 32671792 Free PMC article. Review.
-
Dissecting KMT2D missense mutations in Kabuki syndrome patients.Hum Mol Genet. 2018 Nov 1;27(21):3651-3668. doi: 10.1093/hmg/ddy241. Hum Mol Genet. 2018. PMID: 30107592 Free PMC article.
-
Absence of MSY2 in mouse oocytes perturbs oocyte growth and maturation, RNA stability, and the transcriptome.Biol Reprod. 2011 Sep;85(3):575-83. doi: 10.1095/biolreprod.111.091710. Epub 2011 May 25. Biol Reprod. 2011. PMID: 21613634 Free PMC article.
-
Epigenetic reprogramming during the maternal-to-zygotic transition.MedComm (2020). 2023 Aug 2;4(4):e331. doi: 10.1002/mco2.331. eCollection 2023 Aug. MedComm (2020). 2023. PMID: 37547174 Free PMC article. Review.
References
-
- De La Fuente R, Viveiros M. M, Burns K. H, Adashi E. Y, Matzuk M. M, et al. Major chromatin remodeling in the germinal vesicle (GV) of mammalian oocytes is dispensable for global transcriptional silencing but required for centromeric heterochromatin function. Dev Biol. 2004;275:447–458. - PubMed
-
- Farthing C. R, Ficz G, Ng R. K, Chan C. F, Andrews S, et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pbio.1000116. - DOI - PMC - PubMed
-
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

