Runx2 transcriptome of prostate cancer cells: insights into invasiveness and bone metastasis
- PMID: 20863401
- PMCID: PMC2955618
- DOI: 10.1186/1476-4598-9-258
Runx2 transcriptome of prostate cancer cells: insights into invasiveness and bone metastasis
Abstract
Background: Prostate cancer (PCa) cells preferentially metastasize to bone at least in part by acquiring osteomimetic properties. Runx2, an osteoblast master transcription factor, is aberrantly expressed in PCa cells, and promotes their metastatic phenotype. The transcriptional programs regulated by Runx2 have been extensively studied during osteoblastogenesis, where it activates or represses _target genes in a context-dependent manner. However, little is known about the gene regulatory networks influenced by Runx2 in PCa cells. We therefore investigated genome wide mRNA expression changes in PCa cells in response to Runx2.
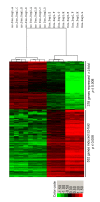
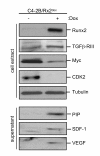
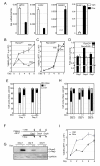
Results: We engineered a C4-2B PCa sub-line called C4-2B/Rx2 dox, in which Doxycycline (Dox) treatment stimulates Runx2 expression from very low to levels observed in other PCa cells. Transcriptome profiling using whole genome expression array followed by in silico analysis indicated that Runx2 upregulated a multitude of genes with prominent cancer associated functions. They included secreted factors (CSF2, SDF-1), proteolytic enzymes (MMP9, CST7), cytoskeleton modulators (SDC2, Twinfilin, SH3PXD2A), intracellular signaling molecules (DUSP1, SPHK1, RASD1) and transcription factors (Sox9, SNAI2, SMAD3) functioning in epithelium to mesenchyme transition (EMT), tissue invasion, as well as homing and attachment to bone. Consistent with the gene expression data, induction of Runx2 in C4-2B cells enhanced their invasiveness. It also promoted cellular quiescence by blocking the G1/S phase transition during cell cycle progression. Furthermore, the cell cycle block was reversed as Runx2 levels declined after Dox withdrawal.
Conclusions: The effects of Runx2 in C4-2B/Rx2 dox cells, as well as similar observations made by employing LNCaP, 22RV1 and PC3 cells, highlight multiple mechanisms by which Runx2 promotes the metastatic phenotype of PCa cells, including tissue invasion, homing to bone and induction of high bone turnover. Runx2 is therefore an attractive _target for the development of novel diagnostic, prognostic and therapeutic approaches to PCa management. _targeting Runx2 may prove more effective than focusing on its individual downstream genes and pathways.
Figures


Similar articles
-
Recruitment of coregulator G9a by Runx2 for selective enhancement or suppression of transcription.J Cell Biochem. 2012 Jul;113(7):2406-14. doi: 10.1002/jcb.24114. J Cell Biochem. 2012. PMID: 22389001 Free PMC article.
-
MicroRNA-466 inhibits tumor growth and bone metastasis in prostate cancer by direct regulation of osteogenic transcription factor RUNX2.Cell Death Dis. 2017 Jan 26;8(1):e2572. doi: 10.1038/cddis.2017.15. Cell Death Dis. 2017. PMID: 28125091 Free PMC article.
-
Role of Runx2 phosphorylation in prostate cancer and association with metastatic disease.Oncogene. 2016 Jan 21;35(3):366-76. doi: 10.1038/onc.2015.91. Epub 2015 Apr 13. Oncogene. 2016. PMID: 25867060 Free PMC article.
-
Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone.Cancer Metastasis Rev. 2006 Dec;25(4):589-600. doi: 10.1007/s10555-006-9032-0. Cancer Metastasis Rev. 2006. PMID: 17165130 Review.
-
Networks and hubs for the transcriptional control of osteoblastogenesis.Rev Endocr Metab Disord. 2006 Jun;7(1-2):1-16. doi: 10.1007/s11154-006-9001-5. Rev Endocr Metab Disord. 2006. PMID: 17051438 Review.
Cited by
-
Epithelial-to-mesenchymal transition in thyroid cancer: a comprehensive review.Endocrine. 2019 Dec;66(3):435-455. doi: 10.1007/s12020-019-02030-8. Epub 2019 Aug 4. Endocrine. 2019. PMID: 31378850 Review.
-
Lung tumor-associated osteoblast-derived bone morphogenetic protein-2 increased epithelial-to-mesenchymal transition of cancer by Runx2/Snail signaling pathway.J Biol Chem. 2011 Oct 28;286(43):37335-46. doi: 10.1074/jbc.M111.256156. Epub 2011 Sep 1. J Biol Chem. 2011. PMID: 21885439 Free PMC article.
-
Ssp1 CaMKK: A Sensor of Actin Polarization That Controls Mitotic Commitment through Srk1 in Schizosaccharomyces pombe.PLoS One. 2015 Nov 17;10(11):e0143037. doi: 10.1371/journal.pone.0143037. eCollection 2015. PLoS One. 2015. PMID: 26575035 Free PMC article.
-
Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine.Mol Cancer. 2020 Dec 12;19(1):171. doi: 10.1186/s12943-020-01293-4. Mol Cancer. 2020. PMID: 33308223 Free PMC article.
-
RUNX2 as a promising therapeutic _target for malignant tumors.Cancer Manag Res. 2021 Mar 16;13:2539-2548. doi: 10.2147/CMAR.S302173. eCollection 2021. Cancer Manag Res. 2021. PMID: 33758548 Free PMC article. Review.
References
-
- Lo Coco F, Pisegna S, Diverio D. The AML1 gene: a transcription factor involved in the pathogenesis of myeloid and lymphoid leukemias. Haematologica. 1997;82:364–370. - PubMed
-
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M. et al._targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/S0092-8674(00)80258-5. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

