Peptide ligands that use a novel binding site to _target both TGF-β receptors
- PMID: 20890540
- PMCID: PMC3064480
- DOI: 10.1039/c0mb00115e
Peptide ligands that use a novel binding site to _target both TGF-β receptors
Abstract
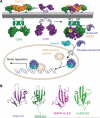
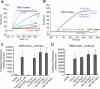
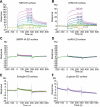
The transforming growth factor beta (TGF-β) signaling pathway plays myriad roles in development and disease. TGF-β isoforms initiate signaling by organizing their cell surface receptors TβRI and TβRII. Exploration and exploitation of the versatility of TGF-β signaling requires an enhanced understanding of structure-function relationships in this pathway. To this end, small molecule, peptide, and antibody effectors that bind key signaling components would serve as valuable probes. We focused on the extracellular domain of TβR1 (TβRI-ED) as a _target for effector screening. The observation that TβRI-ED can bind to a TGF-β coreceptor (endoglin) suggests that the TβRI-ED may have multiple interaction sites. Using phage display, we identified two peptides LTGKNFPMFHRN (Pep1) and MHRMPSFLPTTL (Pep2) that bind the TβRI-ED (K(d)≈ 10(-5) M). Although our screen focused on TβRI-ED, the hit peptides interact with the TβRII-ED with similar affinities. The peptide ligands occupy the same binding sites on TβRI and TβRII, as demonstrated by their ability to compete with each other for receptor binding. Moreover, neither interferes with TGF-β binding. These results indicate that both TβRI and TβRII possess hot spots for protein-protein interactions that are distinct from those used by their known ligand TGF-β. To convert these compounds into high affinity probes, we exploited the observation that TβRI and TβRII exist as dimers on the cell surface; therefore, we assembled a multivalent ligand. Specifically, we displayed one of our receptor-binding peptides on a dendrimer scaffold. We anticipate that the potent multivalent ligand that resulted can be used to probe the role of receptor assembly in TGF-β function.
Figures


Similar articles
-
Assembly of TbetaRI:TbetaRII:TGFbeta ternary complex in vitro with receptor extracellular domains is cooperative and isoform-dependent.J Mol Biol. 2005 Dec 16;354(5):1052-68. doi: 10.1016/j.jmb.2005.10.014. Epub 2005 Oct 27. J Mol Biol. 2005. PMID: 16289576
-
Extracellular and cytoplasmic domains of endoglin interact with the transforming growth factor-beta receptors I and II.J Biol Chem. 2002 Aug 9;277(32):29197-209. doi: 10.1074/jbc.M111991200. Epub 2002 May 15. J Biol Chem. 2002. PMID: 12015308
-
A pivotal role for the transmembrane domain in transforming growth factor-beta receptor activation.J Biol Chem. 1999 Apr 23;274(17):11773-81. doi: 10.1074/jbc.274.17.11773. J Biol Chem. 1999. PMID: 10206994
-
Ternary complex of transforming growth factor-beta1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily.J Biol Chem. 2010 May 7;285(19):14806-14. doi: 10.1074/jbc.M109.079921. Epub 2010 Mar 5. J Biol Chem. 2010. PMID: 20207738 Free PMC article.
-
Regulation of transforming growth factor-beta signaling.Mol Cell Biol Res Commun. 2001 Nov;4(6):321-30. doi: 10.1006/mcbr.2001.0301. Mol Cell Biol Res Commun. 2001. PMID: 11703090 Review.
Cited by
-
Inflammation, Fibrosis and Cancer: Mechanisms, Therapeutic Options and Challenges.Cancers (Basel). 2022 Jan 22;14(3):552. doi: 10.3390/cancers14030552. Cancers (Basel). 2022. PMID: 35158821 Free PMC article. Review.
-
In Vivo Translation of Peptide-_targeted Drug Delivery Systems Discovered by Phage Display.Bioconjug Chem. 2018 Jul 18;29(7):2161-2169. doi: 10.1021/acs.bioconjchem.8b00285. Epub 2018 Jun 29. Bioconjug Chem. 2018. PMID: 29889510 Free PMC article. Review.
-
Small-molecule-modified surfaces engage cells through the αvβ3 integrin.ACS Chem Biol. 2012 Mar 16;7(3):518-25. doi: 10.1021/cb2004725. Epub 2012 Jan 26. ACS Chem Biol. 2012. PMID: 22201290 Free PMC article.
-
Fluorescent "Turn off-on" Small-Molecule-Monitoring Nanoplatform Based on Dendrimer-like Peptides as Competitors.ACS Appl Mater Interfaces. 2019 Sep 11;11(36):33380-33389. doi: 10.1021/acsami.9b13111. Epub 2019 Aug 30. ACS Appl Mater Interfaces. 2019. PMID: 31433617 Free PMC article.
-
TGF-β-Based Therapies for Treating Ocular Surface Disorders.Cells. 2024 Jun 26;13(13):1105. doi: 10.3390/cells13131105. Cells. 2024. PMID: 38994958 Free PMC article. Review.
References
-
- Hinck AP, Archer SJ, Qian SW, Roberts AB, Sporn MB, Weatherbee JA, Tsang MLS, Lucas R, Zhang BL, Wenker J, Torchia DA. Biochemistry. 1996;35:8517–8534. - PubMed
- Mittl PRE, Priestle JP, Cox DA, McMaster G, Cerletti N, Grutter MG. Protein Sci. 1996;5:1261–1271. - PMC - PubMed
- Shi YG, Massague J. Cell. 2003;113:685–700. - PubMed
-
- Hart PJ, Deep S, Taylor AB, Shu ZY, Hinck CS, Hinck AP. Nat. Struct. Biol. 2002;9:203–208. - PubMed
-
- Massague J. Annu. Rev. Biochem. 1998;67:753–791. - PubMed
-
- Deep S, Walker KP, Shu ZY, Hinck AP. Biochemistry. 2003;42:10126–10139. - PubMed
- Boesen CC, Radaev S, Motyka SA, Patamawenu A, Sun PD. Structure. 2002;10:913–919. - PubMed
- Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Nature. 1994;370:341–347. - PubMed
- Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang XF, Massague J. Cell. 1992;71:1003–1014. - PubMed
-
- De Crescenzo G, Hinck CS, Shu ZY, Zuniga J, Yang JH, Tang YP, Baardsnes J, Mendoza V, Sun LZ, Lopez-Casillas F, O'Connor-McCourt M, Hinck AP. J. Mol. Biol. 2006;355:47–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

