A nucleoporin, Nup60p, affects the nuclear and cytoplasmic localization of ASH1 mRNA in S. cerevisiae
- PMID: 21036941
- PMCID: PMC3004054
- DOI: 10.1261/rna.1210411
A nucleoporin, Nup60p, affects the nuclear and cytoplasmic localization of ASH1 mRNA in S. cerevisiae
Abstract
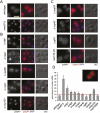
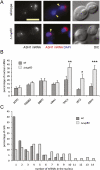
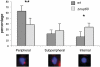
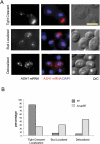
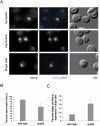
The biogenesis of a localization-competent mRNP begins in the nucleus. It is thought that the coordinated action of nuclear and cytoplasmic components of the localization machinery is required for the efficient export and subsequent subcellular localization of these mRNAs in the cytoplasm. Using quantitative poly(A)(+) and transcript-specific fluorescent in situ hybridization, we analyzed different nonessential nucleoporins and nuclear pore-associated proteins for their potential role in mRNA export and localization. We found that Nup60p, a nuclear pore protein located on the nucleoplasmic side of the nuclear pore complex, was required for the mRNA localization pathway. In a Δnup60 background, localized mRNAs were preferentially retained within the nucleus compared to nonlocalized transcripts. However, the export block was only partial and some transcripts could still reach the cytoplasm. Importantly, downstream processes were also affected. Localization of ASH1 and IST2 mRNAs to the bud was impaired in the Δnup60 background, suggesting that the assembly of a localization competent mRNP ("locasome") was inhibited when NUP60 was deleted. These results demonstrate transcript specificity of a nuclear mRNA retention defect and identify a specific nucleoporin as a functional component of the localization pathway in budding yeast.
Figures
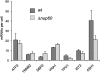
Similar articles
-
Uncoupling of the hnRNP Npl3p from mRNAs during the stress-induced block in mRNA export.Genes Dev. 1999 Aug 1;13(15):1994-2004. doi: 10.1101/gad.13.15.1994. Genes Dev. 1999. PMID: 10444597 Free PMC article.
-
Nucleoporin FG domains facilitate mRNP remodeling at the cytoplasmic face of the nuclear pore complex.Genetics. 2014 Aug;197(4):1213-24. doi: 10.1534/genetics.114.164012. Epub 2014 Jun 14. Genetics. 2014. PMID: 24931410 Free PMC article.
-
Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA.EMBO Rep. 2008 Aug;9(8):781-7. doi: 10.1038/embor.2008.112. Epub 2008 Jun 20. EMBO Rep. 2008. PMID: 18566598 Free PMC article.
-
Principles of mRNA transport in yeast.Cell Mol Life Sci. 2012 Jun;69(11):1843-53. doi: 10.1007/s00018-011-0902-4. Epub 2011 Dec 13. Cell Mol Life Sci. 2012. PMID: 22159587 Free PMC article. Review.
-
Of social molecules: The interactive assembly of ASH1 mRNA-transport complexes in yeast.RNA Biol. 2014;11(8):998-1009. doi: 10.4161/rna.29946. Epub 2014 Oct 31. RNA Biol. 2014. PMID: 25482892 Free PMC article. Review.
Cited by
-
Nuclear mRNA metabolism drives selective basket assembly on a subset of nuclear pore complexes in budding yeast.Mol Cell. 2022 Oct 20;82(20):3856-3871.e6. doi: 10.1016/j.molcel.2022.09.019. Epub 2022 Oct 10. Mol Cell. 2022. PMID: 36220102 Free PMC article.
-
The Hog1 stress-activated protein kinase _targets nucleoporins to control mRNA export upon stress.J Biol Chem. 2013 Jun 14;288(24):17384-98. doi: 10.1074/jbc.M112.444042. Epub 2013 May 3. J Biol Chem. 2013. PMID: 23645671 Free PMC article.
-
Transcriptional elongation and mRNA export are coregulated processes.Genet Res Int. 2011;2011:652461. doi: 10.4061/2011/652461. Epub 2011 Oct 5. Genet Res Int. 2011. PMID: 22567364 Free PMC article.
-
Keeping mRNPs in check during assembly and nuclear export.Nat Rev Mol Cell Biol. 2011 Jun;12(6):377-84. doi: 10.1038/nrm3119. Nat Rev Mol Cell Biol. 2011. PMID: 21602906
-
A subset of FG-nucleoporins is necessary for efficient Msn5-mediated nuclear protein export.Biochim Biophys Acta. 2013 May;1833(5):1096-103. doi: 10.1016/j.bbamcr.2012.12.020. Epub 2013 Jan 4. Biochim Biophys Acta. 2013. PMID: 23295456 Free PMC article.
References
-
- Beach DL, Salmon ED, Bloom K 1999. Localization and anchoring of mRNA in budding yeast. Curr Biol 9: 569–578 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
