Modification of nonstructural protein 1 of influenza A virus by SUMO1
- PMID: 21047957
- PMCID: PMC3020006
- DOI: 10.1128/JVI.00877-10
Modification of nonstructural protein 1 of influenza A virus by SUMO1
Abstract
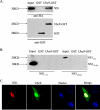
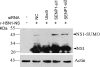
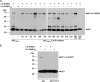
Nonstructural protein 1 (NS1) is one of the major factors resulting in the efficient infection rate and high level of virulence of influenza A virus. Although consisting of only approximately 230 amino acids, NS1 has the ability to interfere with several systems of the host viral defense. In the present study, we demonstrate that NS1 of the highly pathogenic avian influenza A/Duck/Hubei/L-1/2004 (H5N1) virus interacts with human Ubc9, which is the E2 conjugating enzyme for sumoylation, and we show that SUMO1 is conjugated to H5N1 NS1 in both transfected and infected cells. Furthermore, two lysine residues in the C terminus of NS1 were identified as SUMO1 acceptor sites. When the SUMO1 acceptor sites were removed by mutation, NS1 underwent rapid degradation. Studies of different influenza A virus strains of human and avian origin showed that the majority of viruses possess an NS1 protein that is modified by SUMO1, except for the recently emerged swine-origin influenza A virus (S-OIV) (H1N1). Interestingly, growth of a sumoylation-deficient WSN virus mutant was retarded compared to that of wild-type virus. Together, these results indicate that sumoylation enhances NS1 stability and thus promotes rapid growth of influenza A virus.
Figures




Similar articles
-
A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity.Proc Natl Acad Sci U S A. 2008 Mar 18;105(11):4381-6. doi: 10.1073/pnas.0800482105. Epub 2008 Mar 11. Proc Natl Acad Sci U S A. 2008. PMID: 18334632 Free PMC article.
-
A novel SUMOylation site in the influenza a virus NS1 protein identified with a highly sensitive FRET assay.J Biotechnol. 2020 Nov 10;323:121-127. doi: 10.1016/j.jbiotec.2020.08.009. Epub 2020 Aug 19. J Biotechnol. 2020. PMID: 32822681
-
A Conserved Residue, Tyrosine (Y) 84, in H5N1 Influenza A Virus NS1 Regulates IFN Signaling Responses to Enhance Viral Infection.Viruses. 2017 May 12;9(5):107. doi: 10.3390/v9050107. Viruses. 2017. PMID: 28498306 Free PMC article.
-
Virulence determinants of avian H5N1 influenza A virus in mammalian and avian hosts: role of the C-terminal ESEV motif in the viral NS1 protein.J Virol. 2010 Oct;84(20):10708-18. doi: 10.1128/JVI.00610-10. Epub 2010 Aug 4. J Virol. 2010. PMID: 20686040 Free PMC article.
-
The role of nuclear NS1 protein in highly pathogenic H5N1 influenza viruses.Microbes Infect. 2017 Dec;19(12):587-596. doi: 10.1016/j.micinf.2017.08.011. Epub 2017 Sep 10. Microbes Infect. 2017. PMID: 28903072
Cited by
-
TRIM28-mediated nucleocapsid protein SUMOylation enhances SARS-CoV-2 virulence.Nat Commun. 2024 Jan 4;15(1):244. doi: 10.1038/s41467-023-44502-6. Nat Commun. 2024. PMID: 38172120 Free PMC article.
-
Molecular Evolution of the Influenza A Virus Non-structural Protein 1 in Interspecies Transmission and Adaptation.Front Microbiol. 2021 Oct 4;12:693204. doi: 10.3389/fmicb.2021.693204. eCollection 2021. Front Microbiol. 2021. PMID: 34671321 Free PMC article. Review.
-
Complex Virus-Host Interactions Involved in the Regulation of Classical Swine Fever Virus Replication: A Minireview.Viruses. 2017 Jul 5;9(7):171. doi: 10.3390/v9070171. Viruses. 2017. PMID: 28678154 Free PMC article. Review.
-
Viral manipulation of cellular protein conjugation pathways: The SUMO lesson.World J Virol. 2013 May 12;2(2):79-90. doi: 10.5501/wjv.v2.i2.79. World J Virol. 2013. PMID: 24175232 Free PMC article. Review.
-
Rotavirus viroplasm proteins interact with the cellular SUMOylation system: implications for viroplasm-like structure formation.J Virol. 2013 Jan;87(2):807-17. doi: 10.1128/JVI.01578-12. Epub 2012 Oct 31. J Virol. 2013. PMID: 23115286 Free PMC article.
References
-
- Bailey, D., and P. O'Hare. 2002. Herpes simplex virus 1 ICP0 co-localizes with a SUMO-specific protease. J. Gen. Virol. 83:2951-2964. - PubMed
-
- Basler, C. F., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. Garcia-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. U. S. A. 98:2746-2751. - PMC - PubMed
-
- Boggio, R., R. Colombo, R. T. Hay, G. F. Draetta, and S. Chiocca. 2004. A mechanism for inhibiting the SUMO pathway. Mol. Cell 16:549-561. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

