International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid receptors and their ligands: beyond CB₁ and CB₂
- PMID: 21079038
- PMCID: PMC2993256
- DOI: 10.1124/pr.110.003004
International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid receptors and their ligands: beyond CB₁ and CB₂
Abstract
There are at least two types of cannabinoid receptors (CB(1) and CB(2)). Ligands activating these G protein-coupled receptors (GPCRs) include the phytocannabinoid Δ(9)-tetrahydrocannabinol, numerous synthetic compounds, and endogenous compounds known as endocannabinoids. Cannabinoid receptor antagonists have also been developed. Some of these ligands activate or block one type of cannabinoid receptor more potently than the other type. This review summarizes current data indicating the extent to which cannabinoid receptor ligands undergo orthosteric or allosteric interactions with non-CB(1), non-CB(2) established GPCRs, deorphanized receptors such as GPR55, ligand-gated ion channels, transient receptor potential (TRP) channels, and other ion channels or peroxisome proliferator-activated nuclear receptors. From these data, it is clear that some ligands that interact similarly with CB(1) and/or CB(2) receptors are likely to display significantly different pharmacological profiles. The review also lists some criteria that any novel "CB(3)" cannabinoid receptor or channel should fulfil and concludes that these criteria are not currently met by any non-CB(1), non-CB(2) pharmacological receptor or channel. However, it does identify certain pharmacological _targets that should be investigated further as potential CB(3) receptors or channels. These include TRP vanilloid 1, which possibly functions as an ionotropic cannabinoid receptor under physiological and/or pathological conditions, and some deorphanized GPCRs. Also discussed are 1) the ability of CB(1) receptors to form heteromeric complexes with certain other GPCRs, 2) phylogenetic relationships that exist between CB(1)/CB(2) receptors and other GPCRs, 3) evidence for the existence of several as-yet-uncharacterized non-CB(1), non-CB(2) cannabinoid receptors; and 4) current cannabinoid receptor nomenclature.
Figures
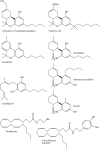
Similar articles
-
Endocannabinoids and Their Pharmacological Actions.Handb Exp Pharmacol. 2015;231:1-37. doi: 10.1007/978-3-319-20825-1_1. Handb Exp Pharmacol. 2015. PMID: 26408156 Review.
-
Receptors and channels _targeted by synthetic cannabinoid receptor agonists and antagonists.Curr Med Chem. 2010;17(14):1360-81. doi: 10.2174/092986710790980050. Curr Med Chem. 2010. PMID: 20166927 Free PMC article. Review.
-
The therapeutic potential of novel cannabinoid receptors.Pharmacol Ther. 2009 May;122(2):83-96. doi: 10.1016/j.pharmthera.2009.01.005. Epub 2009 Feb 25. Pharmacol Ther. 2009. PMID: 19248809 Free PMC article. Review.
-
The pharmacology of cannabinoid receptors and their ligands: an overview.Int J Obes (Lond). 2006 Apr;30 Suppl 1:S13-8. doi: 10.1038/sj.ijo.0803272. Int J Obes (Lond). 2006. PMID: 16570099 Review.
-
WIN55212-2 attenuates amyloid-beta-induced neuroinflammation in rats through activation of cannabinoid receptors and PPAR-γ pathway.Neuropharmacology. 2012 Sep;63(4):653-66. doi: 10.1016/j.neuropharm.2012.05.013. Epub 2012 May 23. Neuropharmacology. 2012. PMID: 22634229
Cited by
-
Lipoxin A4 is an allosteric endocannabinoid that strengthens anandamide-induced CB1 receptor activation.Proc Natl Acad Sci U S A. 2012 Dec 18;109(51):20781-2. doi: 10.1073/pnas.1218529110. Epub 2012 Nov 30. Proc Natl Acad Sci U S A. 2012. PMID: 23204441 Free PMC article. No abstract available.
-
Fatty acid amide hydrolase activity in the dorsal periaqueductal gray attenuates neuropathic pain and associated dysautonomia.Am J Physiol Regul Integr Comp Physiol. 2022 Nov 1;323(5):R749-R762. doi: 10.1152/ajpregu.00073.2022. Epub 2022 Sep 26. Am J Physiol Regul Integr Comp Physiol. 2022. PMID: 36154489 Free PMC article.
-
Microarray and pathway analysis reveal distinct mechanisms underlying cannabinoid-mediated modulation of LPS-induced activation of BV-2 microglial cells.PLoS One. 2013 Apr 24;8(4):e61462. doi: 10.1371/journal.pone.0061462. Print 2013. PLoS One. 2013. PMID: 23637839 Free PMC article.
-
Human metabolites of synthetic cannabinoids JWH-018 and JWH-073 bind with high affinity and act as potent agonists at cannabinoid type-2 receptors.Toxicol Appl Pharmacol. 2013 Jun 1;269(2):100-8. doi: 10.1016/j.taap.2013.03.012. Epub 2013 Mar 26. Toxicol Appl Pharmacol. 2013. PMID: 23537664 Free PMC article.
-
Computational and Experimental Drug Repurposing of FDA-Approved Compounds _targeting the Cannabinoid Receptor CB1.Pharmaceuticals (Basel). 2023 Dec 2;16(12):1678. doi: 10.3390/ph16121678. Pharmaceuticals (Basel). 2023. PMID: 38139805 Free PMC article.
References
-
- Abadji V, Lin S, Taha G, Griffin G, Stevenson LA, Pertwee RG, Makriyannis A. (1994) (R)-methanandamide: a chiral novel anandamide possessing higher potency and metabolic stability. J Med Chem 37:1889–1893 - PubMed
-
- Adam L, Salois D, Rihakova L, Lapointe S, St-Onge S, Labrecque J, Payza K. (2007) Positive allosteric modulators of CB1 receptors, in 17th Annual Symposium of the Cannabinoids; 2007 Jun 26–Jul 1; St-Sauveur, Canada p. 86 International Cannabinoid Research Society, Burlington, Vermont
-
- Ahluwalia J, Urban L, Bevan S, Nagy I. (2003a) Anandamide regulates neuropeptide release from capsaicin-sensitive primary sensory neurons by activating both the cannabinoid 1 receptor and the vanilloid receptor 1 in vitro. Eur J Neurosci 17:2611–2618 - PubMed
-
- Ahluwalia J, Yaqoob M, Urban L, Bevan S, Nagy I. (2003b) Activation of capsaicin-sensitive primary sensory neurones induces anandamide production and release. J Neurochem 84:585–591 - PubMed
-
- Akiyama TE, Sakai S, Lambert G, Nicol CJ, Matsusue K, Pimprale S, Lee YH, Ricote M, Glass CK, Brewer HB, Jr, et al. (2002) Conditional disruption of the peroxisome proliferator-activated receptor gamma gene in mice results in lowered expression of ABCA1, ABCG1, and apoE in macrophages and reduced cholesterol efflux. Mol Cell Biol 22:2607–2619 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DA023204/DA/NIDA NIH HHS/United States
- P50 DA005274/DA/NIDA NIH HHS/United States
- R01 DA003672/DA/NIDA NIH HHS/United States
- DA03672/DA/NIDA NIH HHS/United States
- P01 DA009789/DA/NIDA NIH HHS/United States
- R01 DA023204/DA/NIDA NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01 DA003690/DA/NIDA NIH HHS/United States
- DA03934/DA/NIDA NIH HHS/United States
- R01 DA003934/DA/NIDA NIH HHS/United States
- DA009789/DA/NIDA NIH HHS/United States
- DA021696/DA/NIDA NIH HHS/United States
- DA05274/DA/NIDA NIH HHS/United States
- R37 DA003672/DA/NIDA NIH HHS/United States
- R01 DA003672-27/DA/NIDA NIH HHS/United States
- K05 DA021696/DA/NIDA NIH HHS/United States
- DA03690/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
