Plasma profiles in active systemic juvenile idiopathic arthritis: Biomarkers and biological implications
- PMID: 21136595
- PMCID: PMC3517169
- DOI: 10.1002/pmic.201000298
Plasma profiles in active systemic juvenile idiopathic arthritis: Biomarkers and biological implications
Abstract
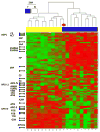
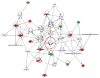
Systemic juvenile idiopathic arthritis (SJIA) is a chronic arthritis of children characterized by a combination of arthritis and systemic inflammation. There is usually non-specific laboratory evidence of inflammation at diagnosis but no diagnostic test. Normalized volumes from 89/889 2-D protein spots representing 26 proteins revealed a plasma pattern that distinguishes SJIA flare from quiescence. Highly discriminating spots derived from 15 proteins constitute a robust SJIA flare signature and show specificity for SJIA flare in comparison to active polyarticular juvenile idiopathic arthritis or acute febrile illness. We used 7 available ELISA assays, including one to the complex of S100A8/S100A9, to measure levels of 8 of the15 proteins. Validating our DIGE results, this ELISA panel correctly classified independent SJIA flare samples, and distinguished them from acute febrile illness. Notably, data using the panel suggest its ability to improve on erythrocyte sedimentation rate or C-reactive protein or S100A8/S100A9, either alone or in combination in SJIA F/Q discriminations. Our results also support the panel's potential clinical utility as a predictor of incipient flare (within 9 wk) in SJIA subjects with clinically inactive disease. Pathway analyses of the 15 proteins in the SJIA flare versus quiescence signature corroborate growing evidence for a key role for IL-1 at disease flare.
Copyright © 2010 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures




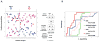
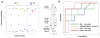
Similar articles
-
Review of biomarkers in systemic juvenile idiopathic arthritis: helpful tools or just playing tricks?Arthritis Res Ther. 2016 Jul 13;18:163. doi: 10.1186/s13075-016-1069-z. Arthritis Res Ther. 2016. PMID: 27411444 Free PMC article. Review.
-
Comparison of biomarkers for systemic juvenile idiopathic arthritis.Pediatr Res. 2015 Nov;78(5):554-9. doi: 10.1038/pr.2015.144. Epub 2015 Aug 12. Pediatr Res. 2015. PMID: 26267155
-
The use of S100 proteins testing in juvenile idiopathic arthritis and autoinflammatory diseases in a pediatric clinical setting: a retrospective analysis.Pediatr Rheumatol Online J. 2020 Jan 16;18(1):7. doi: 10.1186/s12969-020-0398-2. Pediatr Rheumatol Online J. 2020. PMID: 31948488 Free PMC article.
-
Distribution of circulating cells in systemic juvenile idiopathic arthritis across disease activity states.Clin Immunol. 2010 Feb;134(2):206-16. doi: 10.1016/j.clim.2009.09.010. Epub 2009 Oct 29. Clin Immunol. 2010. PMID: 19879195 Free PMC article.
-
Systemic Juvenile Idiopathic Arthritis: Diagnosis and Management.Indian J Pediatr. 2016 Apr;83(4):322-7. doi: 10.1007/s12098-016-2060-z. Epub 2016 Feb 26. Indian J Pediatr. 2016. PMID: 26916892 Review.
Cited by
-
Urine Peptidomic and _targeted Plasma Protein Analyses in the Diagnosis and Monitoring of Systemic Juvenile Idiopathic Arthritis.Clin Proteomics. 2010 Dec;6(4):175-193. doi: 10.1007/s12014-010-9058-8. Epub 2010 Sep 30. Clin Proteomics. 2010. PMID: 21124648 Free PMC article.
-
Monocyte MicroRNA Expression in Active Systemic Juvenile Idiopathic Arthritis Implicates MicroRNA-125a-5p in Polarized Monocyte Phenotypes.Arthritis Rheumatol. 2016 Sep;68(9):2300-13. doi: 10.1002/art.39694. Arthritis Rheumatol. 2016. PMID: 27014994 Free PMC article.
-
Correlation analyses of clinical and molecular findings identify candidate biological pathways in systemic juvenile idiopathic arthritis.BMC Med. 2012 Oct 23;10:125. doi: 10.1186/1741-7015-10-125. BMC Med. 2012. PMID: 23092393 Free PMC article.
-
Synovial membrane protein expression differs between juvenile idiopathic arthritis subtypes in early disease.Arthritis Res Ther. 2014 Jan 13;16(1):R8. doi: 10.1186/ar4434. Arthritis Res Ther. 2014. PMID: 24410838 Free PMC article.
-
Review of biomarkers in systemic juvenile idiopathic arthritis: helpful tools or just playing tricks?Arthritis Res Ther. 2016 Jul 13;18:163. doi: 10.1186/s13075-016-1069-z. Arthritis Res Ther. 2016. PMID: 27411444 Free PMC article. Review.
References
-
- Weiss JE, Ilowite NT. Juvenile idiopathic arthritis. Pediatr Clin North Am. 2005;52:413–442. vi. - PubMed
-
- Singh-Grewal D, Schneider R, Bayer N, Feldman BM. Predictors of disease course and remission in systemic juvenile idiopathic arthritis: significance of early clinical and laboratory features. Arthritis Rheum. 2006;54:1595–1601. - PubMed
-
- Schneider R, Laxer RM. Systemic onset juvenile rheumatoid arthritis. Bailliere’s clinical rheumatology. 1998;12:245–271. - PubMed
-
- Wallace CA, Levinson JE. Juvenile rheumatoid arthritis: outcome and treatment for the 1990s. Rheumatic diseases clinics of North America. 1991;17:891–905. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

