Microarray analysis and barcoded pyrosequencing provide consistent microbial profiles depending on the source of human intestinal samples
- PMID: 21257804
- PMCID: PMC3067328
- DOI: 10.1128/AEM.02477-10
Microarray analysis and barcoded pyrosequencing provide consistent microbial profiles depending on the source of human intestinal samples
Abstract
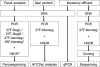
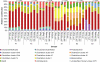
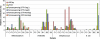
Large-scale and in-depth characterization of the intestinal microbiota necessitates application of high-throughput 16S rRNA gene-based technologies, such as barcoded pyrosequencing and phylogenetic microarray analysis. In this study, the two techniques were compared and contrasted for analysis of the bacterial composition in three fecal and three small intestinal samples from human individuals. As PCR remains a crucial step in sample preparation for both techniques, different forward primers were used for amplification to assess their impact on microbial profiling results. An average of 7,944 pyrosequences, spanning the V1 and V2 region of 16S rRNA genes, was obtained per sample. Although primer choice in barcoded pyrosequencing did not affect species richness and diversity estimates, detection of Actinobacteria strongly depended on the selected primer. Microbial profiles obtained by pyrosequencing and phylogenetic microarray analysis (HITChip) correlated strongly for fecal and ileal lumen samples but were less concordant for ileostomy effluent. Quantitative PCR was employed to investigate the deviations in profiling between pyrosequencing and HITChip analysis. Since cloning and sequencing of random 16S rRNA genes from ileostomy effluent confirmed the presence of novel intestinal phylotypes detected by pyrosequencing, especially those belonging to the Veillonella group, the divergence between pyrosequencing and the HITChip is likely due to the relatively low number of available 16S rRNA gene sequences of small intestinal origin in the DNA databases that were used for HITChip probe design. Overall, this study demonstrated that equivalent biological conclusions are obtained by high-throughput profiling of microbial communities, independent of technology or primer choice.
Figures
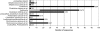
Similar articles
-
Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine.PLoS One. 2009 Aug 20;4(8):e6669. doi: 10.1371/journal.pone.0006669. PLoS One. 2009. PMID: 19693277 Free PMC article.
-
Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison.PLoS One. 2011;6(7):e22788. doi: 10.1371/journal.pone.0022788. Epub 2011 Jul 29. PLoS One. 2011. PMID: 21829515 Free PMC article.
-
Comparative analysis of Korean human gut microbiota by barcoded pyrosequencing.PLoS One. 2011;6(7):e22109. doi: 10.1371/journal.pone.0022109. Epub 2011 Jul 29. PLoS One. 2011. PMID: 21829445 Free PMC article.
-
Application of phylogenetic microarrays to interrogation of human microbiota.FEMS Microbiol Ecol. 2012 Jan;79(1):2-11. doi: 10.1111/j.1574-6941.2011.01222.x. Epub 2011 Nov 9. FEMS Microbiol Ecol. 2012. PMID: 22092522 Free PMC article. Review.
-
Experimental and analytical tools for studying the human microbiome.Nat Rev Genet. 2011 Dec 16;13(1):47-58. doi: 10.1038/nrg3129. Nat Rev Genet. 2011. PMID: 22179717 Free PMC article. Review.
Cited by
-
Microbiota conservation and BMI signatures in adult monozygotic twins.ISME J. 2013 Apr;7(4):707-17. doi: 10.1038/ismej.2012.146. Epub 2012 Nov 29. ISME J. 2013. PMID: 23190729 Free PMC article.
-
Bacterial responses to a simulated colon tumor microenvironment.Mol Cell Proteomics. 2012 Oct;11(10):851-62. doi: 10.1074/mcp.M112.019315. Epub 2012 Jun 19. Mol Cell Proteomics. 2012. PMID: 22713208 Free PMC article.
-
Colonic Mucosal Microbiota and Association of Bacterial Taxa with the Expression of Host Antimicrobial Peptides in Pediatric Ulcerative Colitis.Int J Mol Sci. 2020 Aug 22;21(17):6044. doi: 10.3390/ijms21176044. Int J Mol Sci. 2020. PMID: 32842596 Free PMC article.
-
Application and development of fecal microbiota transplantation in the treatment of gastrointestinal and metabolic diseases: A review.Saudi J Gastroenterol. 2023 Jan-Feb;29(1):3-11. doi: 10.4103/sjg.sjg_131_22. Saudi J Gastroenterol. 2023. PMID: 36412458 Free PMC article. Review.
-
Activity of abundant and rare bacteria in a coastal ocean.Proc Natl Acad Sci U S A. 2011 Aug 2;108(31):12776-81. doi: 10.1073/pnas.1101405108. Epub 2011 Jul 18. Proc Natl Acad Sci U S A. 2011. PMID: 21768380 Free PMC article.
References
-
- Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541-555. - PubMed
-
- Booijink, C. C., et al. 2010. High temporal and inter-individual variation detected in the human ileal microbiota. Environ. Microbiol. 12:3213-3227. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

