Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells
- PMID: 21552279
- PMCID: PMC3124347
- DOI: 10.1038/nature10102
Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells
Abstract
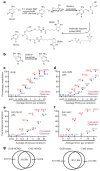
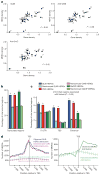
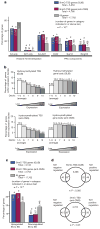
5-hydroxymethylcytosine (5hmC) is a modified base present at low levels in diverse cell types in mammals. 5hmC is generated by the TET family of Fe(II) and 2-oxoglutarate-dependent enzymes through oxidation of 5-methylcytosine (5mC). 5hmC and TET proteins have been implicated in stem cell biology and cancer, but information on the genome-wide distribution of 5hmC is limited. Here we describe two novel and specific approaches to profile the genomic localization of 5hmC. The first approach, termed GLIB (glucosylation, periodate oxidation, biotinylation) uses a combination of enzymatic and chemical steps to isolate DNA fragments containing as few as a single 5hmC. The second approach involves conversion of 5hmC to cytosine 5-methylenesulphonate (CMS) by treatment of genomic DNA with sodium bisulphite, followed by immunoprecipitation of CMS-containing DNA with a specific antiserum to CMS. High-throughput sequencing of 5hmC-containing DNA from mouse embryonic stem (ES) cells showed strong enrichment within exons and near transcriptional start sites. 5hmC was especially enriched at the start sites of genes whose promoters bear dual histone 3 lysine 27 trimethylation (H3K27me3) and histone 3 lysine 4 trimethylation (H3K4me3) marks. Our results indicate that 5hmC has a probable role in transcriptional regulation, and suggest a model in which 5hmC contributes to the 'poised' chromatin signature found at developmentally-regulated genes in ES cells.
Figures
Comment in
-
Epigenetics: Tet proteins in the limelight.Nature. 2011 May 19;473(7347):293-4. doi: 10.1038/473293a. Nature. 2011. PMID: 21593859 No abstract available.
-
Profiling the sixth base.Nat Methods. 2011 Jul;8(7):532-3. doi: 10.1038/nmeth0711-532b. Nat Methods. 2011. PMID: 21850733 No abstract available.
Similar articles
-
Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation.Nature. 2011 May 19;473(7347):398-402. doi: 10.1038/nature10008. Epub 2011 Apr 3. Nature. 2011. PMID: 21460836
-
The GLIB technique for genome-wide mapping of 5-hydroxymethylcytosine.Nat Protoc. 2012 Oct;7(10):1909-17. doi: 10.1038/nprot.2012.104. Epub 2012 Sep 27. Nat Protoc. 2012. PMID: 23018194 Free PMC article.
-
Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase.Genome Biol. 2012 Aug 17;13(8):R69. doi: 10.1186/gb-2012-13-8-r69. Genome Biol. 2012. PMID: 22902005 Free PMC article.
-
Advances in DNA methylation: 5-hydroxymethylcytosine revisited.Clin Chim Acta. 2011 May 12;412(11-12):831-6. doi: 10.1016/j.cca.2011.02.013. Epub 2011 Feb 12. Clin Chim Acta. 2011. PMID: 21324307 Review.
-
TET enzymes and DNA hydroxymethylation in neural development and function - how critical are they?Genomics. 2014 Nov;104(5):334-40. doi: 10.1016/j.ygeno.2014.08.018. Epub 2014 Sep 6. Genomics. 2014. PMID: 25200796 Review.
Cited by
-
The TET2 interactors and their links to hematological malignancies.IUBMB Life. 2015 Jun;67(6):438-45. doi: 10.1002/iub.1389. Epub 2015 Jun 22. IUBMB Life. 2015. PMID: 26099018 Free PMC article. Review.
-
Biochemical characterization of recombinant β-glucosyltransferase and analysis of global 5-hydroxymethylcytosine in unique genomes.Biochemistry. 2012 Feb 7;51(5):1009-19. doi: 10.1021/bi2014739. Epub 2012 Jan 27. Biochemistry. 2012. PMID: 22229759 Free PMC article.
-
Genetic and epigenetic stability of human pluripotent stem cells.Nat Rev Genet. 2012 Oct;13(10):732-44. doi: 10.1038/nrg3271. Epub 2012 Sep 11. Nat Rev Genet. 2012. PMID: 22965355 Review.
-
Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers.Nucleic Acids Res. 2012 Sep 1;40(17):8255-65. doi: 10.1093/nar/gks595. Epub 2012 Jun 22. Nucleic Acids Res. 2012. PMID: 22730288 Free PMC article.
-
Epigenetic Regulation of Neuroinflammation in Alzheimer's Disease.Cells. 2023 Dec 29;13(1):79. doi: 10.3390/cells13010079. Cells. 2023. PMID: 38201283 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- K08 HL089150/HL/NHLBI NIH HHS/United States
- R01 AI044432/AI/NIAID NIH HHS/United States
- R01 HD065812/HD/NICHD NIH HHS/United States
- R01 AI044432-10/AI/NIAID NIH HHS/United States
- K08 HL089150-01A1/HL/NHLBI NIH HHS/United States
- 1 UL1 RR 025758-02/RR/NCRR NIH HHS/United States
- R01 HD065812-01A1/HD/NICHD NIH HHS/United States
- 1 R01 HD065812-01A1/HD/NICHD NIH HHS/United States
- UL1 RR025758/RR/NCRR NIH HHS/United States
- R01 AI44432/AI/NIAID NIH HHS/United States
- RC1 DA028422/DA/NIDA NIH HHS/United States
- RC1 DA028422-02/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

