Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae
- PMID: 21576396
- PMCID: PMC3166859
- DOI: 10.1083/jcb.201102092
Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae
Abstract
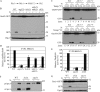
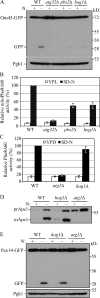
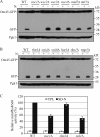
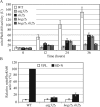
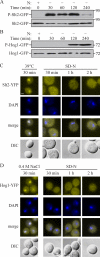
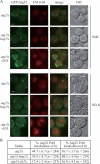
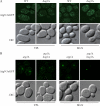
Macroautophagy (hereafter referred to simply as autophagy) is a catabolic pathway that mediates the degradation of long-lived proteins and organelles in eukaryotic cells. The regulation of mitochondrial degradation through autophagy plays an essential role in the maintenance and quality control of this organelle. Compared with our understanding of the essential function of mitochondria in many aspects of cellular metabolism such as energy production and of the role of dysfunctional mitochondria in cell death, little is known regarding their degradation and especially how upstream signaling pathways control this process. Here, we report that two mitogen-activated protein kinases (MAPKs), Slt2 and Hog1, are required for mitophagy in Saccharomyces cerevisiae. Slt2 is required for the degradation of both mitochondria and peroxisomes (via pexophagy), whereas Hog1 functions specifically in mitophagy. Slt2 also affects the recruitment of mitochondria to the phagophore assembly site (PAS), a critical step in the packaging of cargo for selective degradation.
Figures

Similar articles
-
MAPKs regulate mitophagy in Saccharomyces cerevisiae.Autophagy. 2011 Dec;7(12):1564-5. doi: 10.4161/auto.7.12.17971. Autophagy. 2011. PMID: 22024747 Free PMC article.
-
A yeast MAPK cascade regulates pexophagy but not other autophagy pathways.J Cell Biol. 2010 Apr 19;189(2):303-10. doi: 10.1083/jcb.200909154. Epub 2010 Apr 12. J Cell Biol. 2010. PMID: 20385774 Free PMC article.
-
Regulation of the Saccharomyces cerevisiae Slt2 kinase pathway by the stress-inducible Sdp1 dual specificity phosphatase.J Biol Chem. 2002 Jun 14;277(24):21278-84. doi: 10.1074/jbc.M202557200. Epub 2002 Mar 28. J Biol Chem. 2002. PMID: 11923319
-
Pexophagy in yeasts.Biochim Biophys Acta. 2016 May;1863(5):992-8. doi: 10.1016/j.bbamcr.2015.09.023. Epub 2015 Sep 26. Biochim Biophys Acta. 2016. PMID: 26409485 Review.
-
Regulatory mechanisms of mitophagy in yeast.Biochim Biophys Acta Gen Subj. 2021 May;1865(5):129858. doi: 10.1016/j.bbagen.2021.129858. Epub 2021 Feb 3. Biochim Biophys Acta Gen Subj. 2021. PMID: 33545228 Review.
Cited by
-
Atg32-dependent mitophagy sustains spermidine and nitric oxide required for heat-stress tolerance in Saccharomycescerevisiae.J Cell Sci. 2021 Jun 1;134(11):jcs253781. doi: 10.1242/jcs.253781. Epub 2021 Jun 7. J Cell Sci. 2021. PMID: 34096604 Free PMC article.
-
Response to hyperosmotic stress.Genetics. 2012 Oct;192(2):289-318. doi: 10.1534/genetics.112.140863. Genetics. 2012. PMID: 23028184 Free PMC article. Review.
-
The SEACIT complex is involved in the maintenance of vacuole-mitochondria contact sites and controls mitophagy.Cell Mol Life Sci. 2019 Apr;76(8):1623-1640. doi: 10.1007/s00018-019-03015-6. Epub 2019 Jan 23. Cell Mol Life Sci. 2019. PMID: 30673821 Free PMC article.
-
Shedding light on autophagy coordinating with cell wall integrity signaling to govern pathogenicity of Magnaporthe oryzae.Autophagy. 2020 May;16(5):900-916. doi: 10.1080/15548627.2019.1644075. Epub 2019 Jul 24. Autophagy. 2020. PMID: 31313634 Free PMC article.
-
The yeast Saccharomyces cerevisiae: an overview of methods to study autophagy progression.Methods. 2015 Mar;75:3-12. doi: 10.1016/j.ymeth.2014.12.008. Epub 2014 Dec 16. Methods. 2015. PMID: 25526918 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

