Modulation of clock gene expression by the transcriptional coregulator receptor interacting protein 140 (RIP140)
- PMID: 21628546
- PMCID: PMC3207295
- DOI: 10.1177/0748730411401579
Modulation of clock gene expression by the transcriptional coregulator receptor interacting protein 140 (RIP140)
Abstract
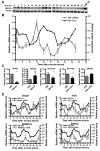
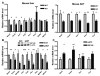
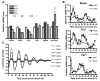
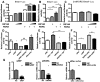
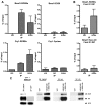
Circadian rhythms are generated in central and peripheral tissues by an intracellular oscillating timing mechanism known as the circadian clock. Several lines of evidence show a strong and bidirectional interplay between metabolism and circadian rhythms. Receptor interacting protein 140 (RIP140) is a coregulator for nuclear receptors and other transcription factors that represses catabolic pathways in metabolic tissues. Although RIP140 functions as a corepressor for most nuclear receptors, mounting evidence points to RIP140 as a dual coregulator that can repress or activate different sets of genes. Here, we demonstrate that RIP140 mRNA and protein levels are under circadian regulation and identify RIP140 as a modulator of clock gene expression, suggesting that RIP140 can participate in a feedback mechanism affecting the circadian clock. We show that the absence of RIP140 disturbs the basal levels of BMAL1 and other clock genes, reducing the amplitude of their oscillations. In addition, we demonstrate that RIP140 is recruited to retinoid-related orphan receptor (ROR) binding sites on the BMAL1 promoter, directly interacts with RORα, and increases transcription from the BMAL1 promoter in a RORα-dependent manner. These results indicate that RIP140 is not only involved in metabolic control but also acts as a coactivator for RORα, influencing clock gene expression.
Figures
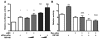
Similar articles
-
Identification of a novel circadian clock modulator controlling BMAL1 expression through a ROR/REV-ERB-response element-dependent mechanism.Biochem Biophys Res Commun. 2016 Jan 15;469(3):580-6. doi: 10.1016/j.bbrc.2015.12.030. Epub 2015 Dec 12. Biochem Biophys Res Commun. 2016. PMID: 26692477
-
The hepatic circadian clock regulates the choline kinase α gene through the BMAL1-REV-ERBα axis.Chronobiol Int. 2015;32(6):774-84. doi: 10.3109/07420528.2015.1046601. Epub 2015 Jun 30. Chronobiol Int. 2015. PMID: 26125130
-
The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1.Nat Struct Mol Biol. 2005 May;12(5):441-8. doi: 10.1038/nsmb925. Epub 2005 Apr 10. Nat Struct Mol Biol. 2005. PMID: 15821743
-
The metabolic coregulator RIP140: an update.Am J Physiol Endocrinol Metab. 2010 Sep;299(3):E335-40. doi: 10.1152/ajpendo.00243.2010. Epub 2010 Jun 8. Am J Physiol Endocrinol Metab. 2010. PMID: 20530738 Review.
-
Control of skeletal muscle metabolic properties by the nuclear receptor corepressor RIP140.Appl Physiol Nutr Metab. 2009 Jun;34(3):362-7. doi: 10.1139/H09-026. Appl Physiol Nutr Metab. 2009. PMID: 19448699 Review.
Cited by
-
Retinoic Acid-Related Orphan Receptors (RORs): Regulatory Functions in Immunity, Development, Circadian Rhythm, and Metabolism.Nucl Receptor Res. 2015;2:101185. doi: 10.11131/2015/101185. Epub 2015 Dec 16. Nucl Receptor Res. 2015. PMID: 26878025 Free PMC article.
-
Expression and Regulatory Network Analysis of miR-140-3p, a New Potential Serum Biomarker for Autism Spectrum Disorder.Front Mol Neurosci. 2017 Aug 10;10:250. doi: 10.3389/fnmol.2017.00250. eCollection 2017. Front Mol Neurosci. 2017. PMID: 28848387 Free PMC article.
-
The PXDLS linear motif regulates circadian rhythmicity through protein-protein interactions.Nucleic Acids Res. 2014 Oct 29;42(19):11879-90. doi: 10.1093/nar/gku873. Epub 2014 Sep 26. Nucleic Acids Res. 2014. PMID: 25260595 Free PMC article.
-
The therapeutic potential of RORγ modulators in the treatment of human disease.J Exp Pharmacol. 2012 Oct 11;4:141-8. doi: 10.2147/JEP.S27078. eCollection 2012. J Exp Pharmacol. 2012. PMID: 27186126 Free PMC article. Review.
-
Nuclear receptors rock around the clock.EMBO Rep. 2014 May;15(5):518-28. doi: 10.1002/embr.201338271. Epub 2014 Apr 15. EMBO Rep. 2014. PMID: 24737872 Free PMC article. Review.
References
-
- Akashi M, Takumi T. (2005) The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol 12:441-448 - PubMed
-
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134:317-328 - PubMed
-
- Augereau P, Badia E, Fuentes M, Rabenoelina F, Corniou M, Derocq D, Balaguer P, Cavailles V. (2006) Transcriptional regulation of the human NRIP1/RIP140 gene by estrogen is modulated by dioxin signalling. Mol Pharmacol 69:1338-1346 - PubMed
-
- Berriel DM, Krones-Herzig A, Metzger D, Ziegler A, Vegiopoulos A, Klingenspor M, Muller-Decker K, Herzig S. (2008) Nuclear receptor cofactor receptor interacting protein 140 controls hepatic triglyceride metabolism during wasting in mice. Hepatology 48:782-791 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

