USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1
- PMID: 21779003
- PMCID: PMC3166460
- DOI: 10.1038/embor.2011.140
USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1
Abstract
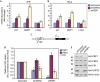
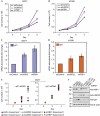
Ubiquitin-specific protease 22 (USP22) edits the histone code by deubiquitinating H2A and H2B as part of the mammalian SAGA (Spt-Ada-Gcn5) complex, and is required for transcriptional regulation and normal cell-cycle progression. Here, we show that USP22 affects the expression of p21 by altering far upstream element (FUSE)-binding protein 1 (FBP1) ubiquitination, as ablation of USP22 leads to increased FBP1 ubiquitination and decreased FBP1 protein occupancy at the p21 gene. Surprisingly, increased polyubiquitination of FBP1 does not alter its protein stability, but instead modulates the stable recruitment of FBP1 to _target loci. Our results indicate a mechanism by which USP22 regulates cell proliferation and tumorigenesis.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


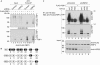
Similar articles
-
Aggregation of Polyglutamine-expanded Ataxin 7 Protein Specifically Sequesters Ubiquitin-specific Protease 22 and Deteriorates Its Deubiquitinating Function in the Spt-Ada-Gcn5-Acetyltransferase (SAGA) Complex.J Biol Chem. 2015 Sep 4;290(36):21996-2004. doi: 10.1074/jbc.M114.631663. Epub 2015 Jul 20. J Biol Chem. 2015. PMID: 26195632 Free PMC article.
-
Ubiquitin-specific protease 22 (USP22) positively regulates RCAN1 protein levels through RCAN1 de-ubiquitination.J Cell Physiol. 2015 Jul;230(7):1651-60. doi: 10.1002/jcp.24917. J Cell Physiol. 2015. PMID: 25546086
-
Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells.J Cell Physiol. 2017 Dec;232(12):3664-3676. doi: 10.1002/jcp.25841. Epub 2017 May 3. J Cell Physiol. 2017. PMID: 28160502
-
Far upstream element binding protein 1: a commander of transcription, translation and beyond.Oncogene. 2013 Jun 13;32(24):2907-16. doi: 10.1038/onc.2012.350. Epub 2012 Aug 27. Oncogene. 2013. PMID: 22926519 Free PMC article. Review.
-
Immune Evasion and Drug Resistance Mediated by USP22 in Cancer: Novel _targets and Mechanisms.Front Immunol. 2022 Jul 20;13:918314. doi: 10.3389/fimmu.2022.918314. eCollection 2022. Front Immunol. 2022. PMID: 35935969 Free PMC article. Review.
Cited by
-
Regulation of pluripotency and differentiation by deubiquitinating enzymes.Cell Death Differ. 2016 Aug;23(8):1257-64. doi: 10.1038/cdd.2016.53. Epub 2016 Jun 10. Cell Death Differ. 2016. PMID: 27285106 Free PMC article. Review.
-
A high-confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex.Mol Cell Biol. 2013 Apr;33(8):1487-502. doi: 10.1128/MCB.00971-12. Epub 2013 Feb 4. Mol Cell Biol. 2013. PMID: 23382074 Free PMC article.
-
The master regulator FUBP1: its emerging role in normal cell function and malignant development.Cell Mol Life Sci. 2019 Jan;76(2):259-281. doi: 10.1007/s00018-018-2933-6. Epub 2018 Oct 20. Cell Mol Life Sci. 2019. PMID: 30343319 Free PMC article. Review.
-
Usp22 Deficiency Leads to Downregulation of PD-L1 and Pathological Activation of CD8+ T Cells and Causes Immunopathology in Response to Acute LCMV Infection.Vaccines (Basel). 2023 Oct 5;11(10):1563. doi: 10.3390/vaccines11101563. Vaccines (Basel). 2023. PMID: 37896966 Free PMC article.
-
Deubiquitinases in cancer.Nat Rev Cancer. 2023 Dec;23(12):842-862. doi: 10.1038/s41568-023-00633-y. Epub 2023 Nov 7. Nat Rev Cancer. 2023. PMID: 37935888 Review.
References
-
- Gartel AL, Radhakrishnan SK (2005) Lost in transcription: p21 epression, mechanisms, and consequences. Cancer Res 65: 3980–3985 - PubMed
-
- Glinsky GV (2006) Genomic models of metastatic cancer: functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathway. Cell Cycle 5: 1208–1216 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

