High-throughput droplet digital PCR system for absolute quantitation of DNA copy number
- PMID: 22035192
- PMCID: PMC3216358
- DOI: 10.1021/ac202028g
High-throughput droplet digital PCR system for absolute quantitation of DNA copy number
Abstract
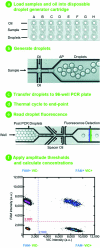
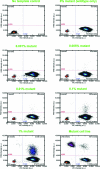
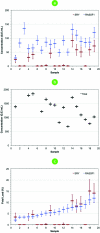
Digital PCR enables the absolute quantitation of nucleic acids in a sample. The lack of scalable and practical technologies for digital PCR implementation has hampered the widespread adoption of this inherently powerful technique. Here we describe a high-throughput droplet digital PCR (ddPCR) system that enables processing of ~2 million PCR reactions using conventional TaqMan assays with a 96-well plate workflow. Three applications demonstrate that the massive partitioning afforded by our ddPCR system provides orders of magnitude more precision and sensitivity than real-time PCR. First, we show the accurate measurement of germline copy number variation. Second, for rare alleles, we show sensitive detection of mutant DNA in a 100,000-fold excess of wildtype background. Third, we demonstrate absolute quantitation of circulating fetal and maternal DNA from cell-free plasma. We anticipate this ddPCR system will allow researchers to explore complex genetic landscapes, discover and validate new disease associations, and define a new era of molecular diagnostics.
Figures

Similar articles
-
Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification.Anal Chem. 2012 Jan 17;84(2):1003-11. doi: 10.1021/ac202578x. Epub 2011 Dec 21. Anal Chem. 2012. PMID: 22122760 Free PMC article.
-
Analyzing Copy Number Variation with Droplet Digital PCR.Methods Mol Biol. 2018;1768:143-160. doi: 10.1007/978-1-4939-7778-9_9. Methods Mol Biol. 2018. PMID: 29717442
-
Quantitative DNA Analysis Using Droplet Digital PCR.Methods Mol Biol. 2017;1492:167-177. doi: 10.1007/978-1-4939-6442-0_11. Methods Mol Biol. 2017. PMID: 27822863
-
Digital Assays Part I: Partitioning Statistics and Digital PCR.SLAS Technol. 2017 Aug;22(4):369-386. doi: 10.1177/2472630317705680. Epub 2017 Apr 27. SLAS Technol. 2017. PMID: 28448765 Review.
-
Droplet digital PCR applications in the tuberculosis world.Tuberculosis (Edinb). 2019 Jul;117:85-92. doi: 10.1016/j.tube.2019.07.001. Epub 2019 Jul 3. Tuberculosis (Edinb). 2019. PMID: 31378274 Review.
Cited by
-
Beyond Tradition: Exploring Cutting-Edge Approaches for Accurate Diagnosis of Human Filariasis.Pathogens. 2024 May 24;13(6):447. doi: 10.3390/pathogens13060447. Pathogens. 2024. PMID: 38921745 Free PMC article. Review.
-
The human ankyrin 1 promoter insulator sustains gene expression in a β-globin lentiviral vector in hematopoietic stem cells.Mol Ther Methods Clin Dev. 2015 Apr 22;2:15012. doi: 10.1038/mtm.2015.12. eCollection 2015. Mol Ther Methods Clin Dev. 2015. PMID: 26029723 Free PMC article.
-
Integrating pathogen- and host-derived blood biomarkers for enhanced tuberculosis diagnosis: a comprehensive review.Front Immunol. 2024 Aug 9;15:1438989. doi: 10.3389/fimmu.2024.1438989. eCollection 2024. Front Immunol. 2024. PMID: 39185416 Free PMC article. Review.
-
Microsatellite Instability in Colorectal Cancer Liquid Biopsy-Current Updates on Its Potential in Non-Invasive Detection, Prognosis and as a Predictive Marker.Diagnostics (Basel). 2021 Mar 18;11(3):544. doi: 10.3390/diagnostics11030544. Diagnostics (Basel). 2021. PMID: 33803882 Free PMC article. Review.
-
Chip in a lab: Microfluidics for next generation life science research.Biomicrofluidics. 2013 Jan;7(1):11302. doi: 10.1063/1.4789751. Epub 2013 Jan 31. Biomicrofluidics. 2013. PMID: 23460772 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

