RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos
- PMID: 22065773
- PMCID: PMC3251083
- DOI: 10.1073/pnas.1112664108
RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos
Abstract
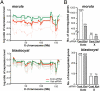
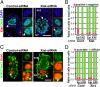
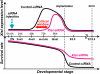
Cloning mammals by somatic cell nuclear transfer (SCNT) is highly inefficient. Most SCNT-generated embryos die after implantation because of unidentified, complex epigenetic errors in the process of postimplantation embryonic development. Here we identify the most upstream level of dysfunction leading to impaired development of clones by using RNAi against Xist, a gene responsible for X chromosome inactivation (XCI). A prior injection of Xist-specific siRNA into reconstructed oocytes efficiently corrected SCNT-specific aberrant Xist expression at the morula stage, but failed to do so thereafter at the blastocyst stage. However, we found that shortly after implantation, this aberrant XCI status in cloned embryos had been corrected autonomously in both embryonic and extraembryonic tissues, probably through a newly established XCI control for postimplantation embryos. Embryo transfer experiments revealed that siRNA-treated embryos showed 10 times higher survival than controls as early as embryonic day 5.5 and this high survival persisted until term, resulting in a remarkable improvement in cloning efficiency (12% vs. 1% in controls). Importantly, unlike control clones, these Xist-siRNA clones at birth showed only a limited dysregulation of their gene expression, indicating that correction of Xist expression in preimplantation embryos had a long-term effect on their postnatal normality. Thus, contrary to the general assumption, our results suggest that the fate of cloned embryos is determined almost exclusively before implantation by their XCI status. Furthermore, our strategy provides a promising breakthrough for mammalian SCNT cloning, because RNAi treatment of oocytes is readily applicable to most mammal species.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
More with less Xist.Proc Natl Acad Sci U S A. 2012 Jan 10;109(2):349-50. doi: 10.1073/pnas.1118081109. Epub 2011 Dec 28. Proc Natl Acad Sci U S A. 2012. PMID: 22205767 Free PMC article. No abstract available.
Similar articles
-
Improvement of developmental competence of cloned male pig embryos by short hairpin ribonucleic acid (shRNA) vector-based but not small interfering RNA (siRNA)-mediated RNA interference (RNAi) of Xist expression.J Reprod Dev. 2019 Dec 18;65(6):533-539. doi: 10.1262/jrd.2019-070. Epub 2019 Oct 21. J Reprod Dev. 2019. PMID: 31631092 Free PMC article.
-
RNAi-mediated knockdown of Xist does not rescue the impaired development of female cloned mouse embryos.J Reprod Dev. 2013;59(3):231-7. doi: 10.1262/jrd.2012-195. Epub 2013 Jan 30. J Reprod Dev. 2013. PMID: 23363561 Free PMC article.
-
RNAi-mediated knockdown of Xist improves development of the female buffalo (Bubalus bubalis) nuclear transfer embryos.Theriogenology. 2022 Jul 15;187:27-33. doi: 10.1016/j.theriogenology.2022.04.020. Epub 2022 Apr 25. Theriogenology. 2022. PMID: 35500424
-
The Role of Xist in X-Chromosome Dosage Compensation.Trends Cell Biol. 2018 Dec;28(12):999-1013. doi: 10.1016/j.tcb.2018.05.005. Epub 2018 Jun 14. Trends Cell Biol. 2018. PMID: 29910081 Free PMC article. Review.
-
X chromosome imprinting and inactivation in the early mammalian embryo.Trends Genet. 1996 Apr;12(4):134-8. doi: 10.1016/0168-9525(96)10017-2. Trends Genet. 1996. PMID: 8901417 Review.
Cited by
-
Insights into epigenetic patterns in mammalian early embryos.Protein Cell. 2021 Jan;12(1):7-28. doi: 10.1007/s13238-020-00757-z. Epub 2020 Jul 15. Protein Cell. 2021. PMID: 32671792 Free PMC article. Review.
-
MicroRNAs in regulation of pluripotency and somatic cell reprogramming: small molecule with big impact.RNA Biol. 2013 Aug;10(8):1255-61. doi: 10.4161/rna.25828. Epub 2013 Jul 23. RNA Biol. 2013. PMID: 23921205 Free PMC article.
-
Improvement of developmental competence of cloned male pig embryos by short hairpin ribonucleic acid (shRNA) vector-based but not small interfering RNA (siRNA)-mediated RNA interference (RNAi) of Xist expression.J Reprod Dev. 2019 Dec 18;65(6):533-539. doi: 10.1262/jrd.2019-070. Epub 2019 Oct 21. J Reprod Dev. 2019. PMID: 31631092 Free PMC article.
-
Development of reproductive engineering techniques at the RIKEN BioResource Center.Exp Anim. 2017 Jan 27;66(1):1-16. doi: 10.1538/expanim.16-0074. Epub 2016 Oct 19. Exp Anim. 2017. PMID: 27760894 Free PMC article. Review.
-
Deletion of the XIST promoter from the human inactive X chromosome compromises polycomb heterochromatin maintenance.Chromosoma. 2021 Sep;130(2-3):177-197. doi: 10.1007/s00412-021-00754-z. Epub 2021 Mar 21. Chromosoma. 2021. PMID: 33745031 Free PMC article.
References
-
- Meissner A, Jaenisch R. Mammalian nuclear transfer. Dev Dyn. 2006;235:2460–2469. - PubMed
-
- Gurdon J, Murdoch A. Nuclear transfer and iPS may work best together. Cell Stem Cell. 2008;2(2):135–138. - PubMed
-
- Thuan NV, Kishigami S, Wakayama T. How to improve the success rate of mouse cloning technology. J Reprod Dev. 2010;56(1):20–30. - PubMed
-
- Okamoto I, Heard E. Lessons from comparative analysis of X-chromosome inactivation in mammals. Chromosome Res. 2009;17:659–669. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

