BRCA1 and BRCA2: different roles in a common pathway of genome protection
- PMID: 22193408
- PMCID: PMC4972490
- DOI: 10.1038/nrc3181
BRCA1 and BRCA2: different roles in a common pathway of genome protection
Abstract
The proteins encoded by the two major breast cancer susceptibility genes, BRCA1 and BRCA2, work in a common pathway of genome protection. However, the two proteins work at different stages in the DNA damage response (DDR) and in DNA repair. BRCA1 is a pleiotropic DDR protein that functions in both checkpoint activation and DNA repair, whereas BRCA2 is a mediator of the core mechanism of homologous recombination. The links between the two proteins are not well understood, but they must exist to explain the marked similarity of human cancer susceptibility that arises with germline mutations in these genes. As discussed here, the proteins work in concert to protect the genome from double-strand DNA damage during DNA replication.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
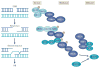
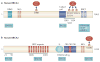

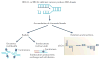
Comment in
-
Initiation, evolution, phenotype and outcome of BRCA1 and BRCA2 mutation-associated breast cancer.Nat Rev Cancer. 2012 May 24;12(5):372-3; author reply 372. doi: 10.1038/nrc3181-c1. Nat Rev Cancer. 2012. PMID: 22525576 No abstract available.
-
BRCA1 and BRCA2: a common pathway of genome protection but different breast cancer subtypes.Nat Rev Cancer. 2012 May 24;12(5):372; author reply 372. doi: 10.1038/nrc3181-c2. Nat Rev Cancer. 2012. PMID: 22525577 No abstract available.
Similar articles
-
PALB2 links BRCA1 and BRCA2 in the DNA-damage response.Curr Biol. 2009 Mar 24;19(6):524-9. doi: 10.1016/j.cub.2009.02.018. Epub 2009 Mar 5. Curr Biol. 2009. PMID: 19268590 Free PMC article.
-
New complexities for BRCA1 and BRCA2.Curr Biol. 2001 Aug 21;11(16):R668-76. doi: 10.1016/s0960-9822(01)00389-x. Curr Biol. 2001. PMID: 11525763 Review.
-
Therapeutic exploitation of tumor cell defects in homologous recombination.Anticancer Agents Med Chem. 2008 May;8(4):448-60. doi: 10.2174/187152008784220267. Anticancer Agents Med Chem. 2008. PMID: 18473729 Review.
-
Hallmarks of 'BRCAness' in sporadic cancers.Nat Rev Cancer. 2004 Oct;4(10):814-9. doi: 10.1038/nrc1457. Nat Rev Cancer. 2004. PMID: 15510162 Review.
-
A BRCA1 Coiled-Coil Domain Variant Disrupting PALB2 Interaction Promotes the Development of Mammary Tumors and Confers a _targetable Defect in Homologous Recombination Repair.Cancer Res. 2021 Dec 15;81(24):6171-6182. doi: 10.1158/0008-5472.CAN-21-1415. Epub 2021 Sep 21. Cancer Res. 2021. PMID: 34548335 Free PMC article.
Cited by
-
Cis-regulatory elements in conserved non-coding sequences of nuclear receptor genes indicate for crosstalk between endocrine systems.Open Med (Wars). 2021 Apr 12;16(1):640-650. doi: 10.1515/med-2021-0264. eCollection 2021. Open Med (Wars). 2021. PMID: 33954257 Free PMC article.
-
The BRCA1 tumor suppressor binds to inositol 1,4,5-trisphosphate receptors to stimulate apoptotic calcium release.J Biol Chem. 2015 Mar 13;290(11):7304-13. doi: 10.1074/jbc.M114.611186. Epub 2015 Feb 2. J Biol Chem. 2015. PMID: 25645916 Free PMC article.
-
Oncogenes induce the cancer-associated fibroblast phenotype: metabolic symbiosis and "fibroblast addiction" are new therapeutic _targets for drug discovery.Cell Cycle. 2013 Sep 1;12(17):2723-32. doi: 10.4161/cc.25695. Epub 2013 Jul 30. Cell Cycle. 2013. PMID: 23860382 Free PMC article. Review.
-
14-weeks combined exercise epigenetically modulated 118 genes of menopausal women with prediabetes.Front Endocrinol (Lausanne). 2022 Aug 15;13:895489. doi: 10.3389/fendo.2022.895489. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36046788 Free PMC article.
-
Human single-stranded DNA binding proteins are essential for maintaining genomic stability.BMC Mol Biol. 2013 Apr 1;14:9. doi: 10.1186/1471-2199-14-9. BMC Mol Biol. 2013. PMID: 23548139 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

