p53 mutants induce transcription of NF-κB2 in H1299 cells through CBP and STAT binding on the NF-κB2 promoter and gain of function activity
- PMID: 22198284
- PMCID: PMC3272778
- DOI: 10.1016/j.abb.2011.12.006
p53 mutants induce transcription of NF-κB2 in H1299 cells through CBP and STAT binding on the NF-κB2 promoter and gain of function activity
Abstract
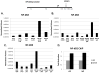
Cancer cells with p53 mutations, in general, grow more aggressively than those with wild-type p53 and show "gain of function" (GOF) phenotypes such as increased growth rate, enhanced resistance to chemotherapeutic drugs, increased cell motility and tumorigenicity; although the mechanism for this function remains unknown. In this communication we report that p53-mediated NF-κB2 up-regulation significantly contributes to the aggressive oncogenic behavior of cancer cells. Lowering the level of mutant p53 in a number of cancer cell lines resulted in a loss of GOF phenotypes directly implicating p53 mutants in the process. RNAi against NF-κB2 in naturally occurring cancer cell lines also lowers GOF activities. In H1299 cells expressing mutant p53, chromatin immunoprecipitation (ChIP) assays indicate that mutant p53 induces histone acetylation at specific sites on the regulatory regions of its _target genes. ChIP assays using antibodies against transcription factors putatively capable of interacting with the NF-κB2 promoter show increased interaction of CBP and STAT2 in the presence of mutant p53. Thus, we propose that in H1299 cells, mutant p53 elevates expression of genes capable of enhancing cell proliferation, motility, and tumorigenicity by inducing acetylation of histones via recruitment of CBP and STAT2 on the promoters causing CBP-mediated histone acetylation.
Copyright © 2011. Published by Elsevier Inc.
Figures






Similar articles
-
Gain-of-function mutant p53 upregulates CXC chemokines and enhances cell migration.Carcinogenesis. 2012 Feb;33(2):442-51. doi: 10.1093/carcin/bgr270. Epub 2011 Nov 22. Carcinogenesis. 2012. PMID: 22114072
-
Tumor-derived p53 mutants induce NF-kappaB2 gene expression.Mol Cell Biol. 2005 Nov;25(22):10097-110. doi: 10.1128/MCB.25.22.10097-10110.2005. Mol Cell Biol. 2005. PMID: 16260623 Free PMC article.
-
Addiction of lung cancer cells to GOF p53 is promoted by up-regulation of epidermal growth factor receptor through multiple contacts with p53 transactivation domain and promoter.Onco_target. 2016 Mar 15;7(11):12426-46. doi: 10.18632/onco_target.6998. Onco_target. 2016. PMID: 26820293 Free PMC article.
-
How do nuclear factor kappa B (NF-κB)1 and NF-κB2 defects lead to the incidence of clinical and immunological manifestations of inborn errors of immunity?Expert Rev Clin Immunol. 2023 Mar;19(3):329-339. doi: 10.1080/1744666X.2023.2174105. Epub 2023 Feb 9. Expert Rev Clin Immunol. 2023. PMID: 36706462 Review.
-
Harnessing the vulnerabilities of p53 mutants in lung cancer - Focusing on the proteasome: a new trick for an old foe?Cancer Biol Ther. 2020 Apr 2;21(4):293-302. doi: 10.1080/15384047.2019.1702403. Epub 2020 Feb 10. Cancer Biol Ther. 2020. PMID: 32041464 Free PMC article. Review.
Cited by
-
p53 mutations in cancer.Nat Cell Biol. 2013 Jan;15(1):2-8. doi: 10.1038/ncb2641. Nat Cell Biol. 2013. PMID: 23263379 Review.
-
Mutant p53 establishes _targetable tumor dependency by promoting unscheduled replication.J Clin Invest. 2017 May 1;127(5):1839-1855. doi: 10.1172/JCI87724. Epub 2017 Apr 10. J Clin Invest. 2017. PMID: 28394262 Free PMC article.
-
Functional expression of mitochondrial KCa3.1 channels in non-small cell lung cancer cells.Pflugers Arch. 2022 Nov;474(11):1147-1157. doi: 10.1007/s00424-022-02748-x. Epub 2022 Sep 24. Pflugers Arch. 2022. PMID: 36152073 Free PMC article.
-
More _targets, more pathways and more clues for mutant p53.Oncogenesis. 2013 Jul 1;2(7):e54. doi: 10.1038/oncsis.2013.15. Oncogenesis. 2013. PMID: 23817466 Free PMC article.
-
Molecular crosstalk between cancer and neurodegenerative diseases.Cell Mol Life Sci. 2020 Jul;77(14):2659-2680. doi: 10.1007/s00018-019-03428-3. Epub 2019 Dec 28. Cell Mol Life Sci. 2020. PMID: 31884567 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

