Balancing repair and tolerance of DNA damage caused by alkylating agents
- PMID: 22237395
- PMCID: PMC3586545
- DOI: 10.1038/nrc3185
Balancing repair and tolerance of DNA damage caused by alkylating agents
Abstract
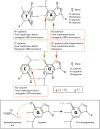
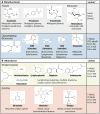
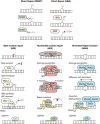
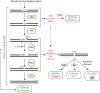
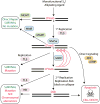
Alkylating agents constitute a major class of frontline chemotherapeutic drugs that inflict cytotoxic DNA damage as their main mode of action, in addition to collateral mutagenic damage. Numerous cellular pathways, including direct DNA damage reversal, base excision repair (BER) and mismatch repair (MMR), respond to alkylation damage to defend against alkylation-induced cell death or mutation. However, maintaining a proper balance of activity both within and between these pathways is crucial for a favourable response of an organism to alkylating agents. Furthermore, the response of an individual to alkylating agents can vary considerably from tissue to tissue and from person to person, pointing to genetic and epigenetic mechanisms that modulate alkylating agent toxicity.
Figures

Similar articles
-
DNA alkylation lesion repair: outcomes and implications in cancer chemotherapy.J Zhejiang Univ Sci B. 2021 Jan 15;22(1):47-62. doi: 10.1631/jzus.B2000344. J Zhejiang Univ Sci B. 2021. PMID: 33448187 Free PMC article. Review.
-
MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents.DNA Repair (Amst). 2007 Aug 1;6(8):1079-99. doi: 10.1016/j.dnarep.2007.03.008. Epub 2007 May 7. DNA Repair (Amst). 2007. PMID: 17485253 Review.
-
Alkylation damage in DNA and RNA--repair mechanisms and medical significance.DNA Repair (Amst). 2004 Nov 2;3(11):1389-407. doi: 10.1016/j.dnarep.2004.05.004. DNA Repair (Amst). 2004. PMID: 15380096 Review.
-
The human cyclin B1 protein modulates sensitivity of DNA mismatch repair deficient prostate cancer cell lines to alkylating agents.Exp Cell Res. 2000 May 25;257(1):127-34. doi: 10.1006/excr.2000.4865. Exp Cell Res. 2000. PMID: 10854060
-
Distinct pathways for repairing mutagenic lesions induced by methylating and ethylating agents.Mutagenesis. 2013 May;28(3):341-50. doi: 10.1093/mutage/get010. Epub 2013 Feb 27. Mutagenesis. 2013. PMID: 23446177 Free PMC article.
Cited by
-
XRCC1 codon 280 polymorphism and susceptibility to lung cancer: a meta-analysis of the literatures.Tumour Biol. 2013 Oct;34(5):2989-94. doi: 10.1007/s13277-013-0863-3. Epub 2013 Jun 4. Tumour Biol. 2013. PMID: 23733202
-
Augmented HR Repair Mediates Acquired Temozolomide Resistance in Glioblastoma.Mol Cancer Res. 2016 Oct;14(10):928-940. doi: 10.1158/1541-7786.MCR-16-0125. Epub 2016 Jun 29. Mol Cancer Res. 2016. PMID: 27358111 Free PMC article.
-
Accidental Encounter of Repair Intermediates in Alkylated DNA May Lead to Double-Strand Breaks in Resting Cells.Int J Mol Sci. 2024 Jul 26;25(15):8192. doi: 10.3390/ijms25158192. Int J Mol Sci. 2024. PMID: 39125763 Free PMC article. Review.
-
Base damage within single-strand DNA underlies in vivo hypermutability induced by a ubiquitous environmental agent.PLoS Genet. 2012;8(12):e1003149. doi: 10.1371/journal.pgen.1003149. Epub 2012 Dec 13. PLoS Genet. 2012. PMID: 23271983 Free PMC article.
-
Past, Present, and Future of Anticancer Nanomedicine.Int J Nanomedicine. 2020 Aug 6;15:5719-5743. doi: 10.2147/IJN.S254774. eCollection 2020. Int J Nanomedicine. 2020. PMID: 32821098 Free PMC article. Review.
References
-
- Ballschmiter K. Pattern and sources of naturally produced organohalogens in the marine environment: biogenic formation of organohalogens. Chemosphere. 2003;52:313–24. - PubMed
-
- Hecht SS. DNA adduct formation from tobacco-specific N-nitrosamines. Mutat Res. 1999;424:127–42. - PubMed
-
- Hamilton JT, McRoberts WC, Keppler F, Kalin RM, Harper DB. Chloride methylation by plant pectin: an efficient environmentally significant process. Science. 2003;301:206–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

