Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin
- PMID: 22280891
- PMCID: PMC3288275
- DOI: 10.1016/j.devcel.2011.12.014
Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin
Abstract
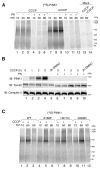
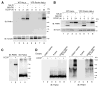
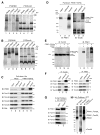
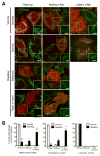
Mutations in the mitochondrial kinase PINK1 and the cytosolic E3 ligase Parkin can cause Parkinson's disease. Damaged mitochondria accumulate PINK1 on the outer membrane where, dependent on kinase activity, it recruits and activates Parkin to induce mitophagy, potentially maintaining organelle fidelity. How PINK1 recruits Parkin is unknown. We show that endogenous PINK1 forms a 700 kDa complex with the translocase of the outer membrane (TOM) selectively on depolarized mitochondria whereas PINK1 ectopically _targeted to the outer membrane retains association with TOM on polarized mitochondria. Inducibly _targeting PINK1 to peroxisomes or lysosomes, which lack a TOM complex, recruits Parkin and activates ubiquitin ligase activity on the respective organelles. Once there, Parkin induces organelle selective autophagy of peroxisomes but not lysosomes. We propose that the association of PINK1 with the TOM complex allows rapid reimport of PINK1 to rescue repolarized mitochondria from mitophagy, and discount mitochondrial-specific factors for Parkin translocation and activation.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Parkin recruitment to impaired mitochondria for nonselective ubiquitylation is facilitated by MITOL.J Biol Chem. 2019 Jun 28;294(26):10300-10314. doi: 10.1074/jbc.RA118.006302. Epub 2019 May 20. J Biol Chem. 2019. PMID: 31110043 Free PMC article.
-
The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy.Nature. 2015 Aug 20;524(7565):309-314. doi: 10.1038/nature14893. Epub 2015 Aug 12. Nature. 2015. PMID: 26266977 Free PMC article.
-
Deciphering the Molecular Signals of PINK1/Parkin Mitophagy.Trends Cell Biol. 2016 Oct;26(10):733-744. doi: 10.1016/j.tcb.2016.05.008. Epub 2016 Jun 10. Trends Cell Biol. 2016. PMID: 27291334 Review.
-
A dimeric PINK1-containing complex on depolarized mitochondria stimulates Parkin recruitment.J Biol Chem. 2013 Dec 20;288(51):36372-84. doi: 10.1074/jbc.M113.509653. Epub 2013 Nov 4. J Biol Chem. 2013. PMID: 24189060 Free PMC article.
-
The Role of PTEN-L in Modulating PINK1-Parkin-Mediated Mitophagy.Neurotox Res. 2022 Aug;40(4):1103-1114. doi: 10.1007/s12640-022-00475-w. Epub 2022 Jun 14. Neurotox Res. 2022. PMID: 35699891 Review.
Cited by
-
Minireview: Autophagy in pancreatic β-cells and its implication in diabetes.Mol Endocrinol. 2015 Mar;29(3):338-48. doi: 10.1210/me.2014-1367. Epub 2015 Jan 29. Mol Endocrinol. 2015. PMID: 25633274 Free PMC article. Review.
-
P2X7 Receptor and Purinergic Signaling: Orchestrating Mitochondrial Dysfunction in Neurodegenerative Diseases.eNeuro. 2022 Nov 14;9(6):ENEURO.0092-22.2022. doi: 10.1523/ENEURO.0092-22.2022. Print 2022 Nov-Dec. eNeuro. 2022. PMID: 36376084 Free PMC article. Review.
-
Mitochondria in Multi-Directional Differentiation of Dental-Derived Mesenchymal Stem Cells.Biomolecules. 2023 Dec 21;14(1):12. doi: 10.3390/biom14010012. Biomolecules. 2023. PMID: 38275753 Free PMC article. Review.
-
Miro phosphorylation sites regulate Parkin recruitment and mitochondrial motility.Proc Natl Acad Sci U S A. 2016 Oct 11;113(41):E6097-E6106. doi: 10.1073/pnas.1612283113. Epub 2016 Sep 27. Proc Natl Acad Sci U S A. 2016. PMID: 27679849 Free PMC article.
-
ATM mediates spermidine-induced mitophagy via PINK1 and Parkin regulation in human fibroblasts.Sci Rep. 2016 Apr 19;6:24700. doi: 10.1038/srep24700. Sci Rep. 2016. PMID: 27089984 Free PMC article.
References
-
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. - PubMed
-
- Brix J, Dietmeier K, Pfanner N. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J Biol Chem. 1997;272:20730–20735. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

